Search Count: 43
 |
Organism: Mus musculus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: MOTOR PROTEIN Ligands: PO4, MG, ADP |
 |
Organism: Mus musculus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: MOTOR PROTEIN Ligands: MG, ADP |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: MOTOR PROTEIN Ligands: PO4, MG, ADP |
 |
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-04-27 Classification: TOXIN |
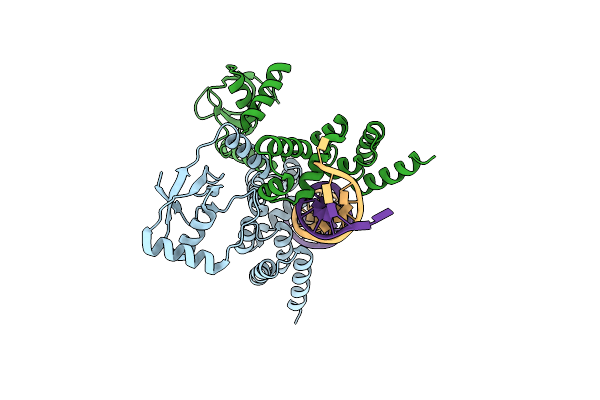 |
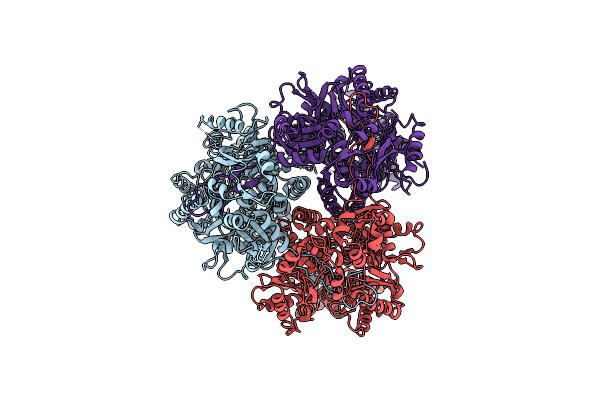
Crystal Structure Of C-Terminally Truncated Bacillus Subtilis Nucleoid Occlusion Protein (Noc) Complexed To The Noc-Binding Site (Nbs)
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-03-09 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: MEMBRANE PROTEIN |
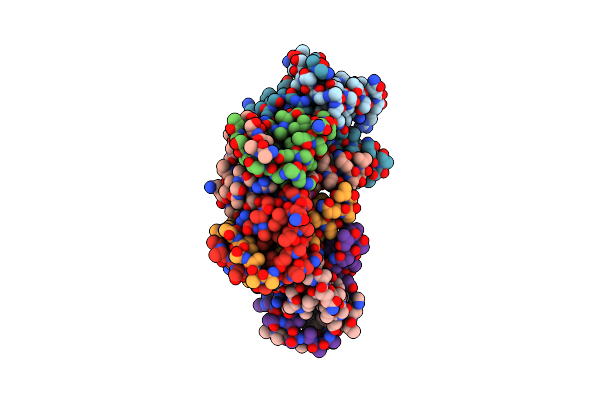 |
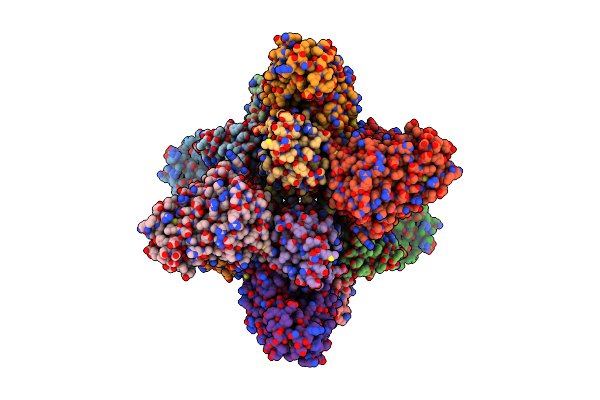
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2021-10-06 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: CELL ADHESION Ligands: PIO |
 |
Crystal Structure Of Human Serum Albumin In Complex With Myristic Acid At 2.27 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2021-02-24 Classification: PROTEIN TRANSPORT Ligands: MYR, MPD, FMT, PO4 |
 |
Crystal Structure Of Human Serum Albumin In Complex With Perfluorooctanoic Acid (Pfoa) At 2.10 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-24 Classification: PROTEIN TRANSPORT Ligands: 8PF, MPO, PGE, PG4, MYR, MPD, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-01-27 Classification: CELL CYCLE |
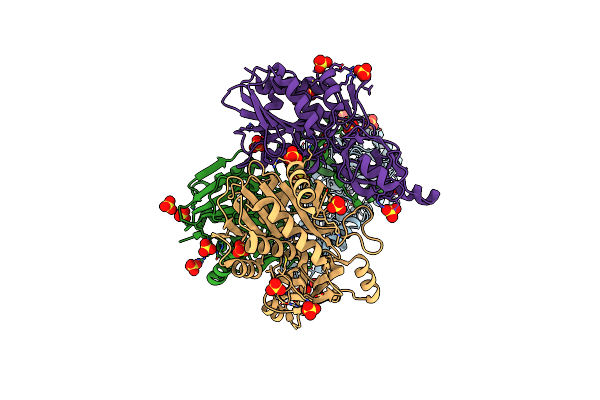 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: ZN, CO, SO4, OH |
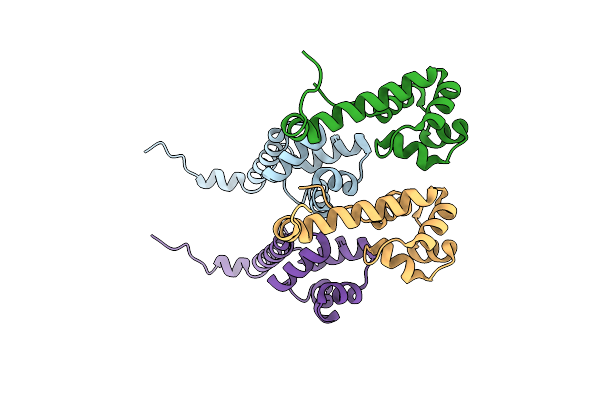 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: ANTITOXIN |
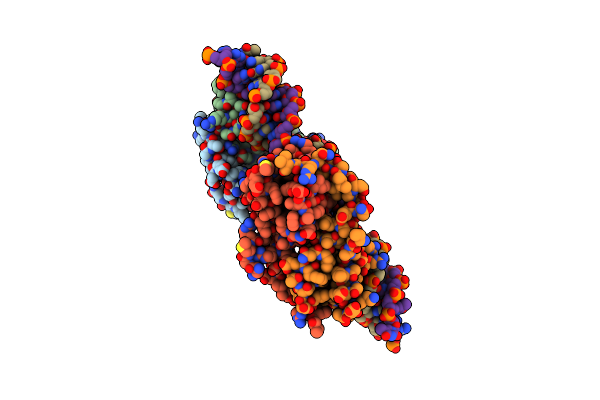 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-01-30 Classification: ANTITOXIN |
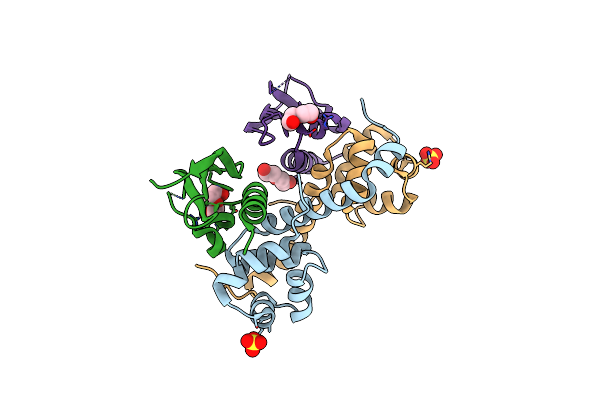 |
Organism: Pseudomonas putida, Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-11 Classification: TOXIN Ligands: SO4, PEG |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-16 Classification: LYASE Ligands: CIT |
 |
Crystal Structure Of H60A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-05-16 Classification: LYASE Ligands: CIT |
 |
Crystal Structure Of H307A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-05-16 Classification: LYASE |
 |
Structure Of Phosphorylated Translation Elongation Factor Ef-Tu From E. Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: MG, GDP |

