Search Count: 29
 |
Nadh:Quinone Oxidoreductase (Ndh-Ii) From Staphylococcus Aureus - Holoprotein Structure - 2.32 A Resolution
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: FAD, PO4, MLI |
 |
Nadh:Quinone Oxidoreductase (Ndh-Ii) From Staphylococcus Aureus - E172S Mutant
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-10-19 Classification: OXIDOREDUCTASE Ligands: FE, OXY, FMN |
 |
Oxidized Flavodiiron Core Of Escherichia Coli Flavorubredoxin, Including The Fe-4Sg Atoms From Its Rubredoxin Domain
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-10-19 Classification: OXIDOREDUCTASE Ligands: FEO, FMN, PO4, CAC, ACY, GOL, FE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: EDO, PGE, PEG, GOL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: PGE, EDO, SO4, ZN, DHL, GOL, PG4, FLC |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-09-03 Classification: ELECTRON TRANSPORT Ligands: FMN, FE, O, PO4, CL, GOL |
 |
The Importance Of The Abn2 Calcium Cluster In The Endo-1,5- Arabinanase Activity From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-03-05 Classification: HYDROLASE Ligands: NI, TRS |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2013-04-10 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, XE |
 |
Structure Of The Tetracycline Degrading Monooxygenase Tetx In Complex With Minocycline
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2012-12-12 Classification: FLAVOPROTEIN Ligands: FAD, MIY, SO4 |
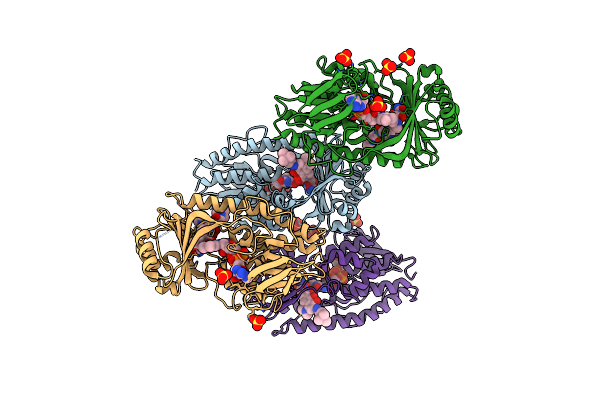 |
Structure Of The Tetracycline Degrading Monooxygenase Tetx In Complex With Tigecycline
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-11-21 Classification: OXIDOREDUCTASE Ligands: FAD, T1C, SO4 |
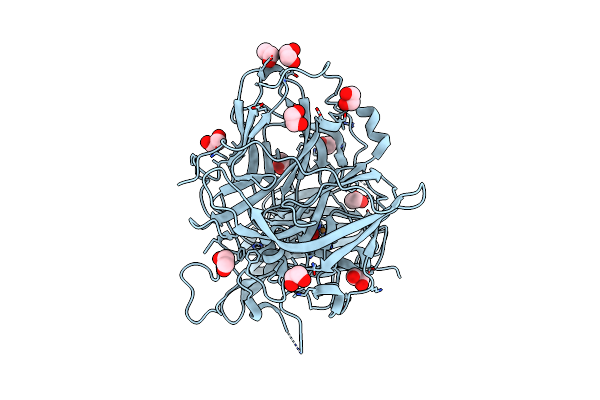 |
Mutations In The Neighbourhood Of Cota-Laccase Trinuclear Site: D116A Mutant
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-01-25 Classification: OXIDOREDUCTASE Ligands: CU, EDO, PER |
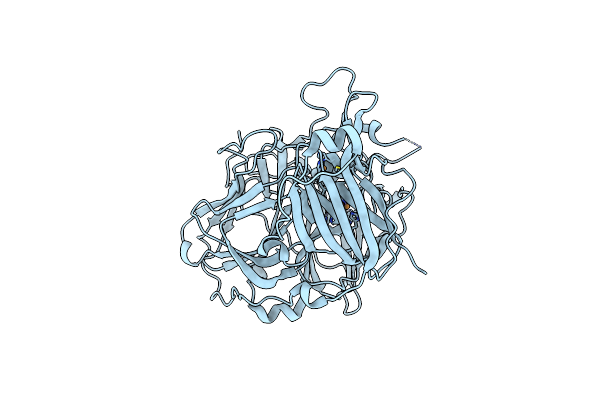 |
Mutations In The Neighbourhood Of Cota-Laccase Trinuclear Site: D116E Mutant
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-25 Classification: OXIDOREDUCTASE Ligands: CU, PER |
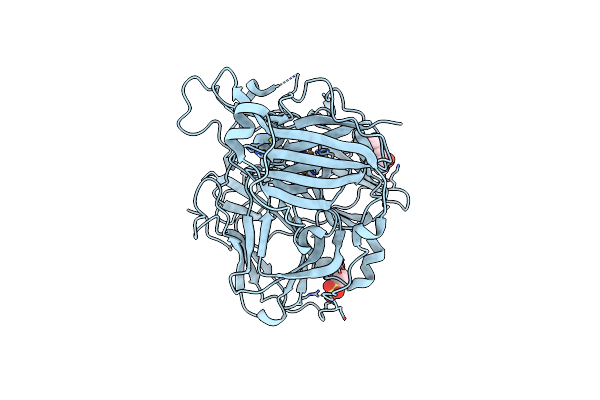 |
Mutations In The Neighbourhood Of Cota-Laccase Trinuclear Site: D116N Mutant
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-25 Classification: OXIDOREDUCTASE Ligands: CU, OH, PER, EPE, MPD |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-09-29 Classification: OXIDOREDUCTASE Ligands: CU, OXY, EDO |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-09-22 Classification: OXIDOREDUCTASE Ligands: CU, OH, EDO |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-22 Classification: OXIDOREDUCTASE Ligands: CU, OXY, MPD |
 |
Mannosyl-3-Phosphoglycerate Synthase From Thermus Thermophilus Hb27 Apoprotein
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: FLC, ZN |
 |
Mannosyl-3-Phosphoglycerate Synthase From Thermus Thermophilus Hb27 In Complex With Gdp-Alpha-D-Mannose And Mg(Ii)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: GDD, MG, ZN |
 |
H309A Mutant Of Mannosyl-3-Phosphoglycerate Synthase From Thermus Thermophilus Hb27 In Complex With Gdp-Alpha-D-Mannose And Mg(Ii)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2010-03-31 Classification: TRANSFERASE Ligands: GDD, MG, ZN |

