Search Count: 114
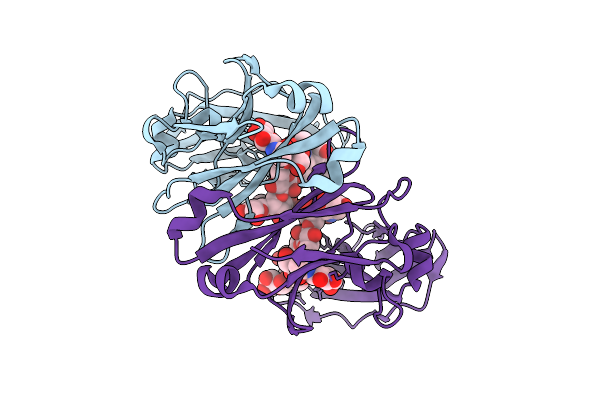 |
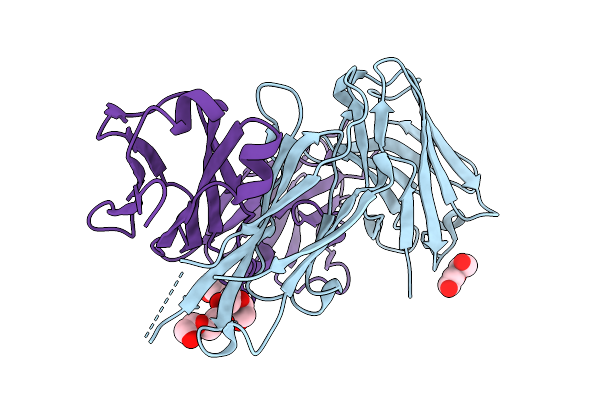
Crystal Structure Of Knob-In-Hole Immunoglobulin G1 Fc Heterodimer With P374A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
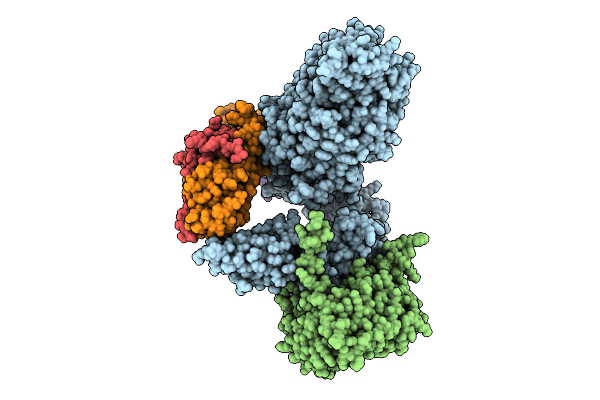 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
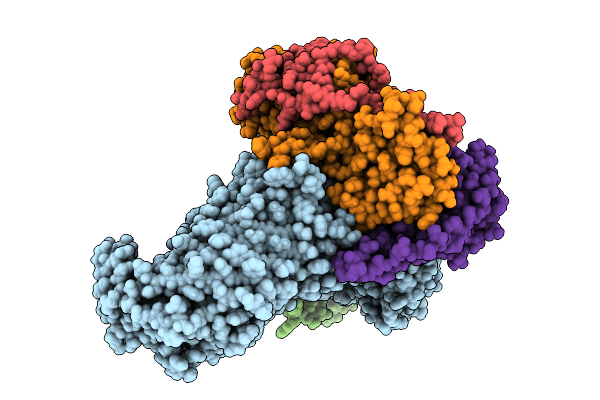 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
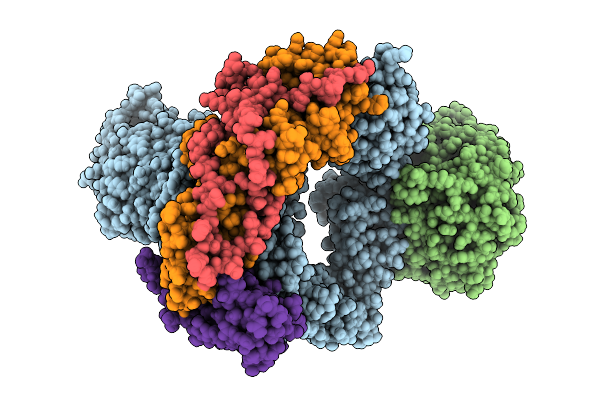 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: PEG |
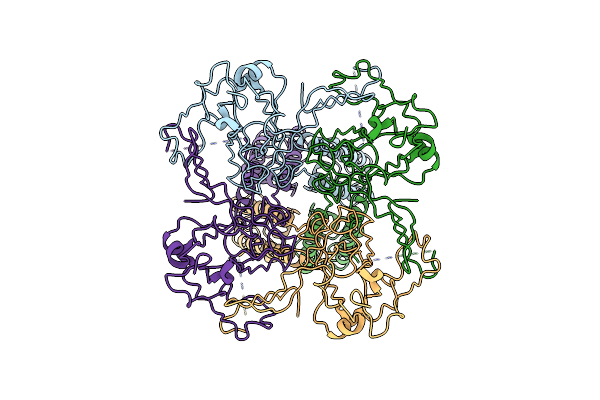 |
Cryo-Em Structure Of The Human Inward-Rectifier Potassium 2.1 Channel (Kir2.1) - R312H Mutant
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN |
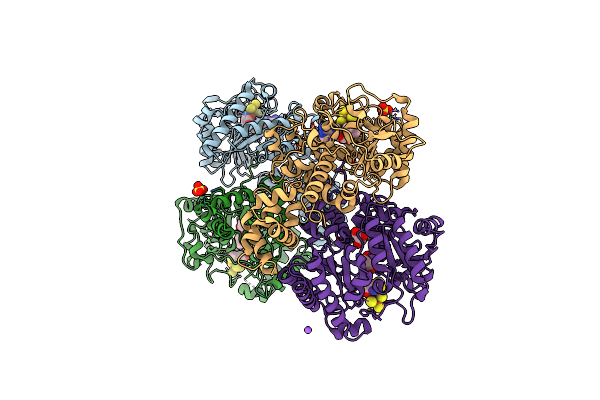 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SAM, MET, SO4, GOL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, MET |
 |
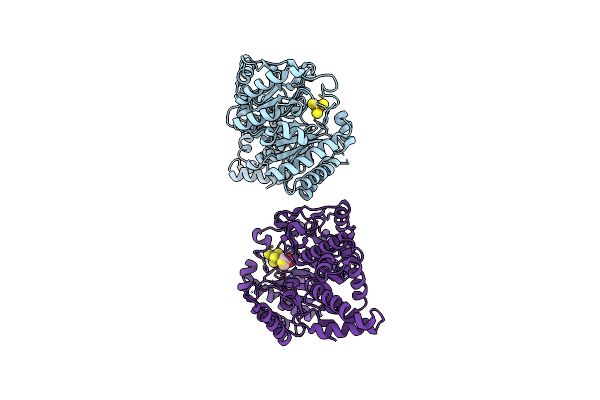
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-411
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHV |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-410
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHU |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-400
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHO |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-409
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHS |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-403
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHW |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-406
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHX, NI |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-1154
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: OBT |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-402
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHY, A1AHZ |
 |
Structural Characterization Of Phox2B And Its Dna Interactions Shed Lights Into The Molecular Basis Of The + 7Ala Variant Pathogenicity In Cchs
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-05-15 Classification: PROTEIN BINDING |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of The Cibeles-Demetra 3:3 Heterocomplex From Galleria Mellonella Saliva
Organism: Galleria mellonella
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: UNKNOWN FUNCTION Ligands: CU |

