Search Count: 22
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: ZN, CA, PO4 |
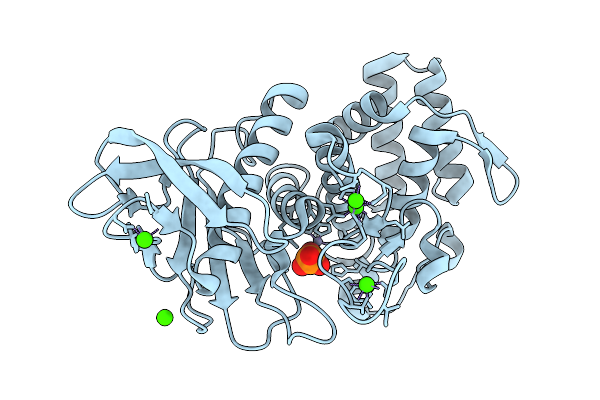 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: ZN, CA, PO4 |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: METAL BINDING PROTEIN Ligands: FE, PEG |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: METAL BINDING PROTEIN Ligands: PEG, SCN |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: PEG, MN |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2024-01-31 Classification: OXIDOREDUCTASE Ligands: PEG |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2024-01-31 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Streptomyces plicatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: HYDROLASE, SUGAR BINDING PROTEIN Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-03-11 Classification: CYTOKINE |
 |
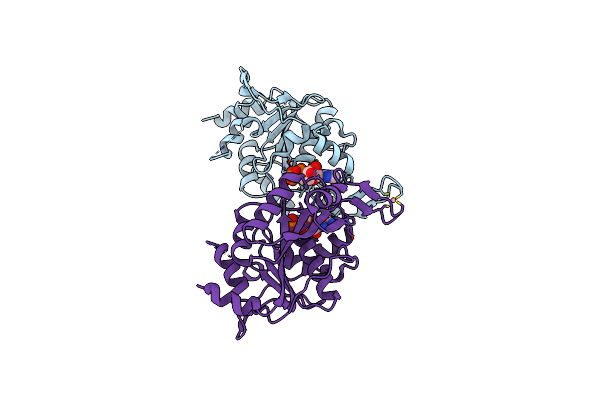
Crystal Structure Of The Putative Histidinol Phosphatase Hisk From Listeria Monocytogenes With Trinuclear Metals Determined By Pixe Revealing Sulphate Ion In Active Site. Based On Pixe Analysis And Original Date From 3Dcp
Organism: Listeria monocytogenes serotype 4b str. h7858
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MN, CO, SO4, FE, CA |
 |
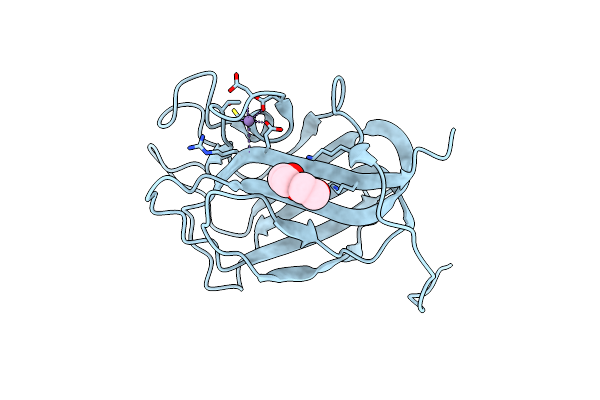
The Nucleotide-Binding Protein Af_226 In Complex With Adp From Archaeoglobus Fulgidus With Co Found By Pixe. Based On 3Kb1.
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2019-12-25 Classification: METAL TRANSPORT Ligands: ADP, CO |
 |
X-Ray Structure Of The C-Terminal Domain (277-440) Of Putative Chitobiase From Bacteroides Thetaiotaomicron. Northeast Structural Genomics Consortium Target Btr324A. Re-Refinement Of 3Ggl With Correct Metal Mn Replacing Zn. New Metal Confirmed With Pixe Analysis Of Original Sample.
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: PEG, MN |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2015-02-04 Classification: HYDROLASE Ligands: SO4, EPE, CL, GOL |

