Search Count: 48
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-03-02 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2022-03-02 Classification: DE NOVO PROTEIN |
 |
Organism: Streptomyces plicatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: HYDROLASE, SUGAR BINDING PROTEIN Ligands: MG |
 |
Ambient-Temperature Serial Femtosecond X-Ray Crystal Structure Of Sars-Cov-2 Main Protease At 1.9 A Resolution (C121)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-30 Classification: VIRAL PROTEIN |
 |
Ambient-Temperature Serial Femtosecond X-Ray Crystal Structure Of Sars-Cov-2 Main Protease At 2.1 A Resolution (P212121)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-09-30 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-03-11 Classification: CYTOKINE |
 |
Crystal Structure Of The Putative Histidinol Phosphatase Hisk From Listeria Monocytogenes With Trinuclear Metals Determined By Pixe Revealing Sulphate Ion In Active Site. Based On Pixe Analysis And Original Date From 3Dcp
Organism: Listeria monocytogenes serotype 4b str. h7858
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MN, CO, SO4, FE, CA |
 |
The Nucleotide-Binding Protein Af_226 In Complex With Adp From Archaeoglobus Fulgidus With Co Found By Pixe. Based On 3Kb1.
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2019-12-25 Classification: METAL TRANSPORT Ligands: ADP, CO |
 |
X-Ray Structure Of The C-Terminal Domain (277-440) Of Putative Chitobiase From Bacteroides Thetaiotaomicron. Northeast Structural Genomics Consortium Target Btr324A. Re-Refinement Of 3Ggl With Correct Metal Mn Replacing Zn. New Metal Confirmed With Pixe Analysis Of Original Sample.
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: PEG, MN |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
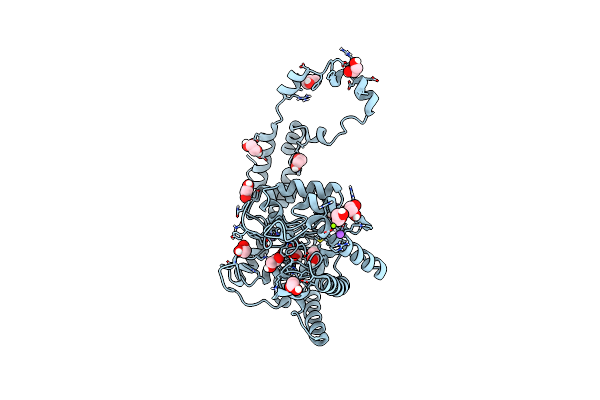 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
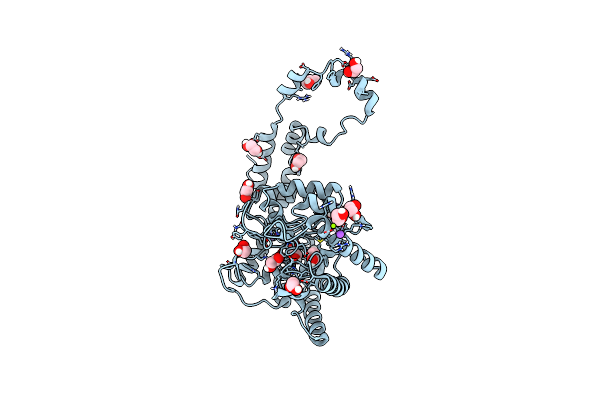 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
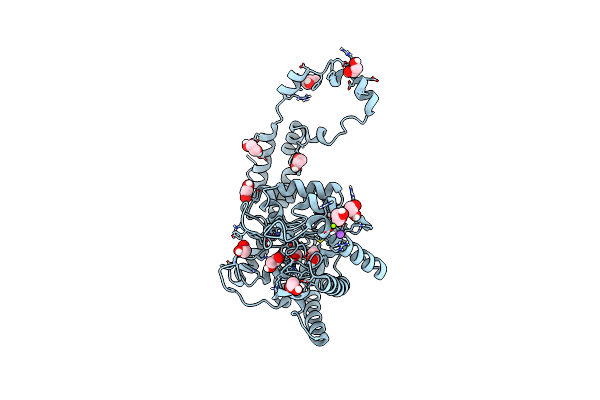 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
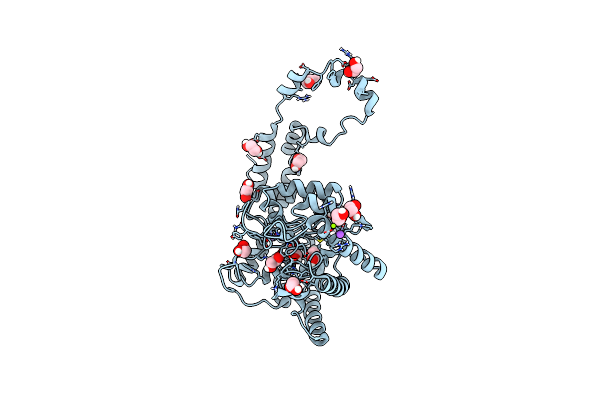 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-07-17 Classification: METAL BINDING PROTEIN Ligands: EDO, IPA, MN, MG, NA |
 |
Cs-Rosetta Determined Structures Of The N-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Cs-Rosetta Determined Structures Of The C-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Direct Visualisation Of Strain-Induced Protein Post-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2015-03-25 Classification: LYASE Ligands: ACO, MG, CL |

