Search Count: 52
 |
Organism: Paulinella chromatophora
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: UNKNOWN FUNCTION Ligands: SO4, CL, EDO |
 |
Organism: Paulinella chromatophora
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: UNKNOWN FUNCTION Ligands: POL |
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley In The Apo State
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley, In Complex With Its Substrate
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
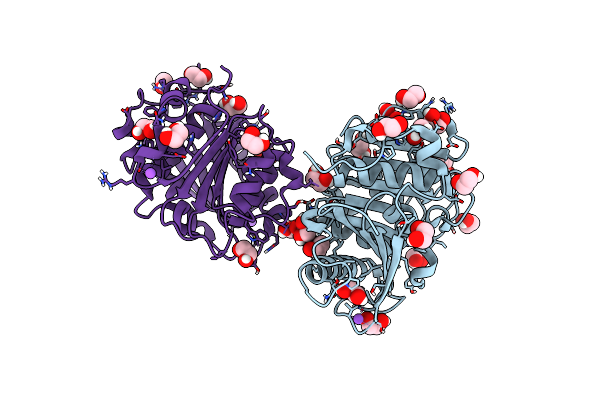
Crystal Structure Of The Tin2-Fold Effector Protein Tue1 From Thecaphora Thlaspeos
Organism: Thecaphora thlaspeos
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: NUCLEAR PROTEIN |
 |
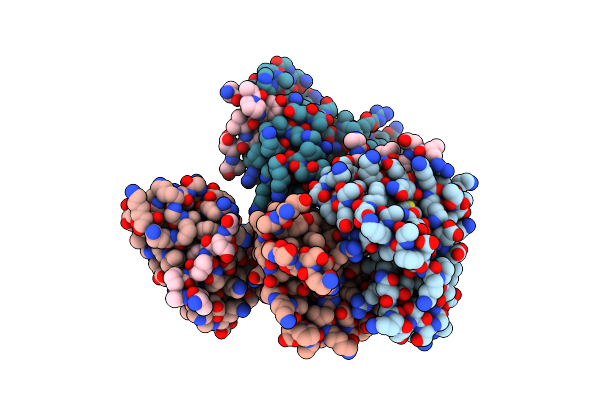
The Crystal Structure Of Pet44, A Petase Enzyme From Alkalilimnicola Ehrlichii
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-11-06 Classification: HYDROLASE |
 |
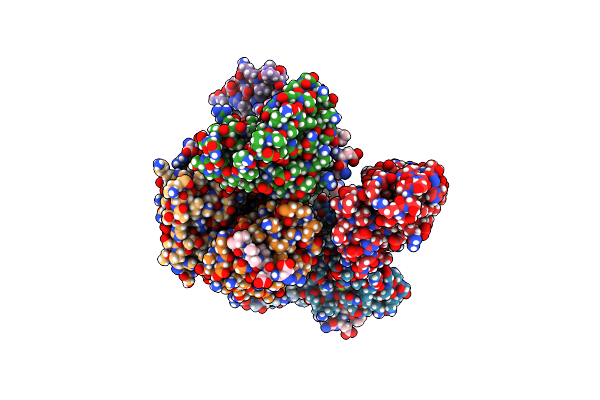
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-10-30 Classification: RNA |
 |
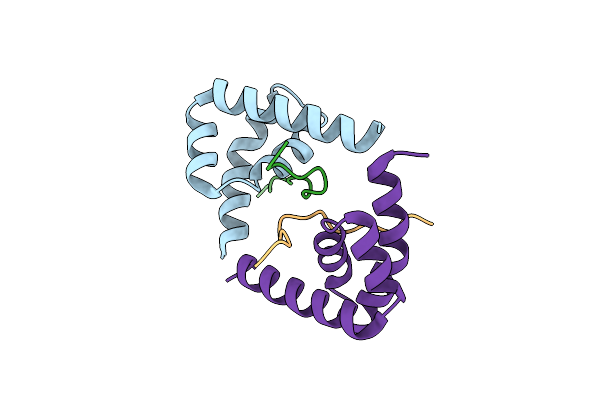
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-10-30 Classification: RNA |
 |
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: RNA BINDING PROTEIN |
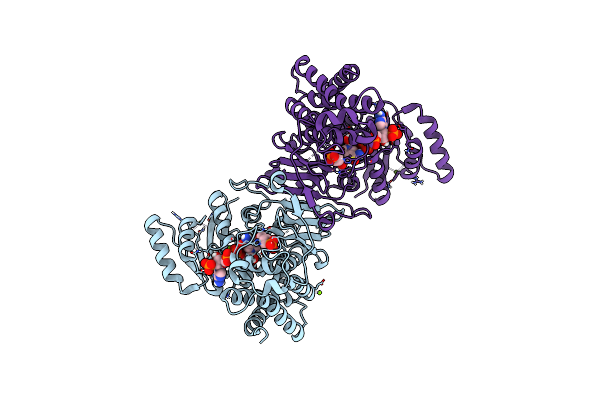 |
1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (Dxr) As Target For Anti Toxoplasma Gondii Compounds: Crystal Structure, Biochemical Characterization And Biological Evaluation Of Inhibitors
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:2.56 Å Release Date: 2024-08-21 Classification: STRUCTURAL PROTEIN Ligands: NAP, FOM, CL, MG |
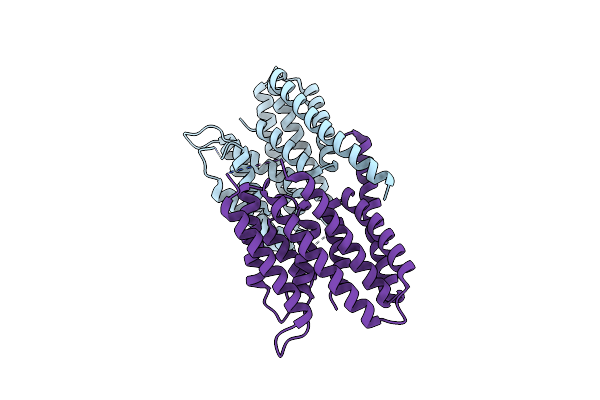 |
Organism: Chlamydia
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-07 Classification: ENDOCYTOSIS |
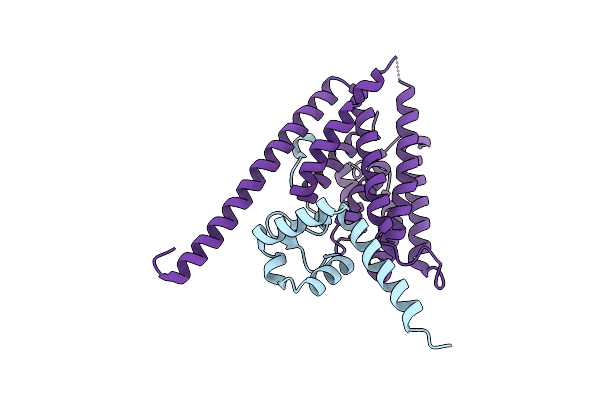 |
Organism: Chlamydia, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-08-07 Classification: ENDOCYTOSIS |
 |
The Crystal Structure Of Pet46, A Petase Enzyme From Candidatus Bathyarchaeota
Organism: Candidatus bathyarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: EDO, PO4, CL |
 |
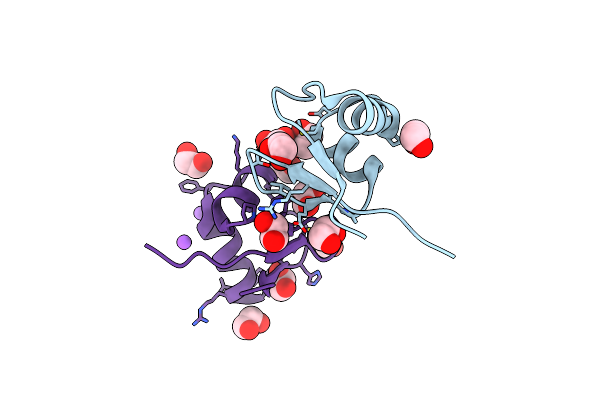
Organism: Acidihalobacter
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: PLP |
 |
Organism: Acidihalobacter
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: GH0 |
 |
Organism: Acidihalobacter
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2023-08-16 Classification: HYDROLASE |
 |
Organism: Acidihalobacter
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-07-26 Classification: HYDROLASE |
 |
Crystal Structure Of The Dna-Binding Short Chromatophore-Targeted Protein Sctp-23166 From Paulinella Chromatophora
Organism: Paulinella chromatophora
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN Ligands: EDO, NA |
 |
The Crystal Structure Of The S178A Mutant Of Pet40, A Petase Enzyme From An Unclassified Amycolatopsis
Organism: Unclassified amycolatopsis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: EPE, EDO, CL, MG |
 |
Organism: Bartonella clarridgeiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-04-05 Classification: TOXIN |

