Search Count: 49
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-10 Classification: HYDROLASE |
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: CA, EDO, BTB |
 |
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: EDO |
 |
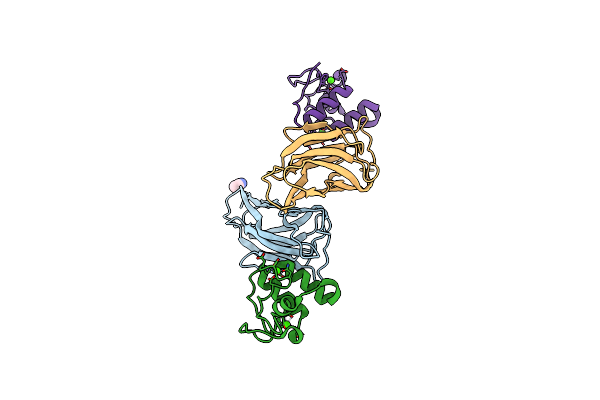
Crystal Structure Of The Kix Domain Of Cbp In Complex With A Mll/C-Myb Chimera
Organism: Homo sapiens, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-09 Classification: TRANSCRIPTION Ligands: GOL, CL |
 |
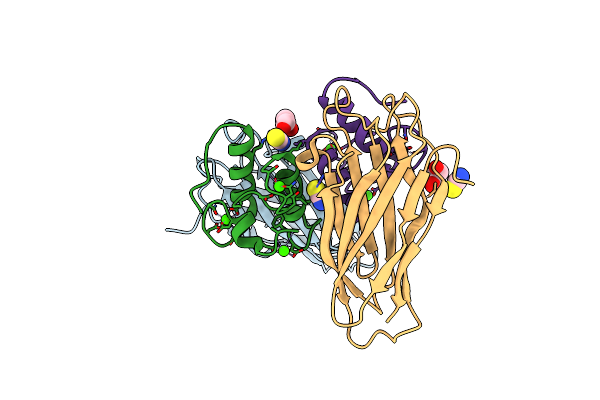
Crystal Structure Of Ruminococcus Flavefaciens' Type Iii Complex Containing The Fifth Cohesin From Scaffoldin B And The Dockerin From Scaffoldin A
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-02-28 Classification: PROTEIN BINDING Ligands: CCN, CA |
 |
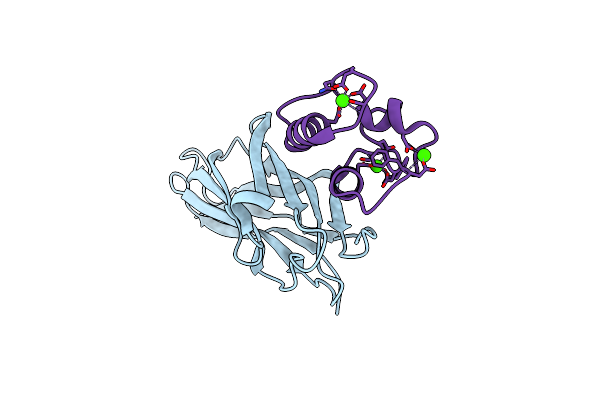
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S15I, I16N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-01-31 Classification: PROTEIN BINDING Ligands: CA, SCN, GOL |
 |
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S51I, L52N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-01-31 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-06 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: SUGAR BINDING PROTEIN Ligands: EDO, BR |
 |
Structural Insight Into Host Cell Surface Retention Of A 1.5-Mda Bacterial Ice-Binding Adhesin
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-19 Classification: CELL ADHESION Ligands: CA, MG, EDO |
 |
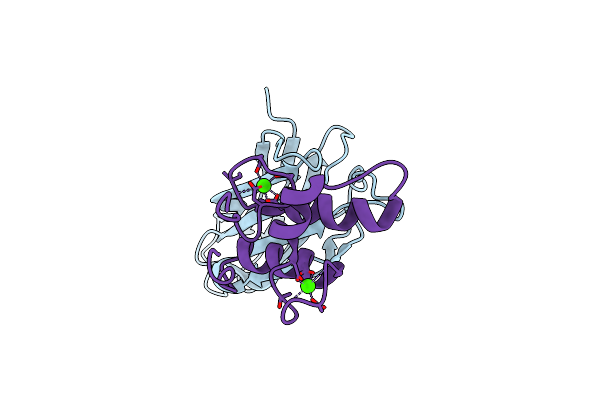
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-07-19 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA |
 |
Organism: Ruminococcus flavefaciens, Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA, GOL |
 |
Organism: Marinomonas primoryensis
Method: SOLUTION NMR Release Date: 2017-06-28 Classification: Antifreeze protein, Cell adhesion |
 |
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2017-06-07 Classification: CELL ADHESION Ligands: CA, GLC, BGC |
 |
Crystal Structure Of Ruminococcus Flavefaciens Scaffoldin C Cohesin In Complex With A Dockerin From An Uncharacterized Cbm-Containing Protein
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-10-19 Classification: PROTEIN BINDING Ligands: CA |
 |
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-08-03 Classification: MOTOR PROTEIN |
 |
Organism: Schizophyllum commune h4-8
Method: SOLUTION NMR Release Date: 2016-03-16 Classification: PROTEIN FIBRIL |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-12-09 Classification: carbohydrate-binding module Ligands: CA, EDO |

