Search Count: 21
 |
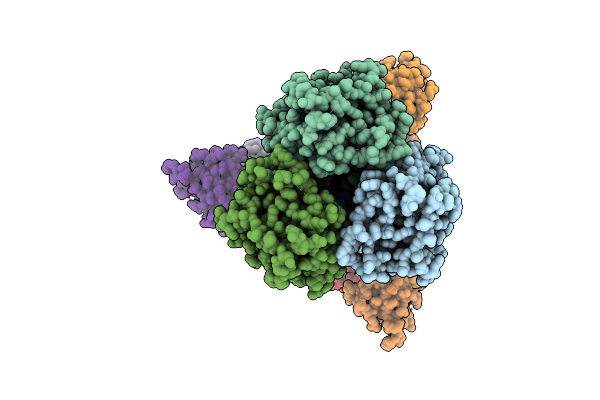
Cryoem Structure Of Nc99 Hemagglutinin Trimer In Complex With Fab Bb798E 3-C07
Organism: Influenza a virus, Macaca fascicularis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN |
 |
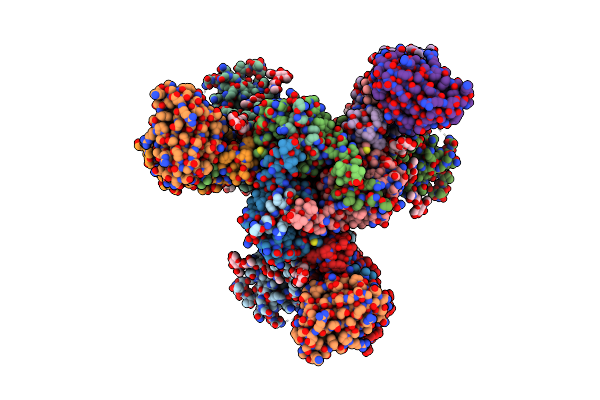
Cryoem Structure Of Nc99 Hemagglutinin Trimer In Complex With Fab T009 3-E04
Organism: Influenza a virus, Macaca fascicularis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN |
 |
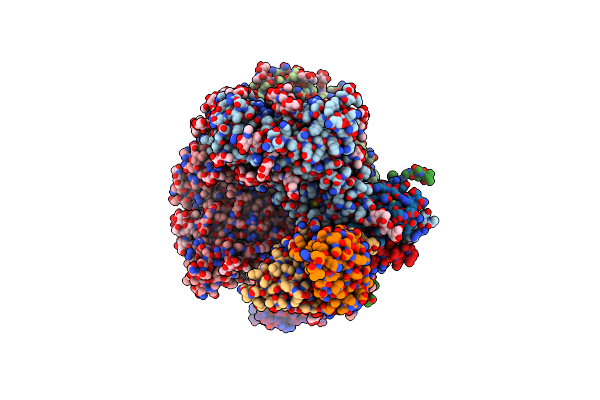
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-B.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
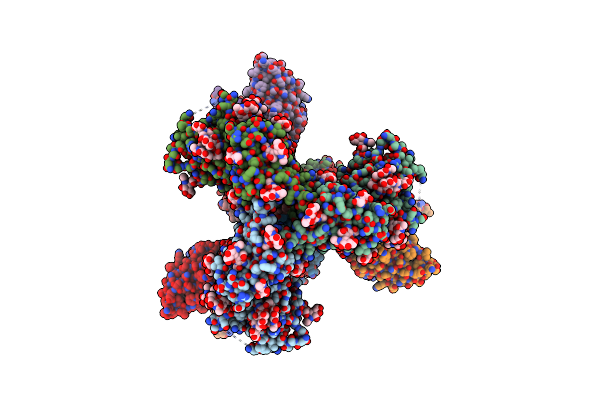
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-D.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-E.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Herh-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-F*01 Fab In Complex With Hiv-1 Env Trimer Conc Sosip
Organism: Homo sapiens, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Env Bg505 Ds-Sosip In Complex With Antibody Gpz6-B.01 Targeting The Fusion Peptide
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-A.01 Fab
Organism: Macaca, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Gpz6-A.01 Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral PROTEIN/iMMUNE SYSTEM Ligands: NAG |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-12-28 Classification: SIGNALING PROTEIN Ligands: NAG, MG |
 |
Metabotropic Glutamate Receptor In Complex With Antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-Azanyl-3-[[3,4-Bis(Fluoranyl)Phenyl]Sulfanylmethyl]-4-Oxidanyl-Bicyclo[3.1.0]Hexane-2,6-Dicarboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-12-28 Classification: SIGNALING PROTEIN Ligands: 6YS, NAG |
 |
Organism: Montipora efflorescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-04-03 Classification: LUMINESCENT PROTEIN Ligands: IOD |
 |
The 2.0 Angstroms Crystal Structure Of A Pocilloporin At Ph 3.5: The Structural Basis For The Linkage Between Color Transition And Halide Binding
Organism: Montipora efflorescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-05 Classification: LUMINESCENT PROTEIN Ligands: IOD, CL, ACY |
 |
The 2.1A Crystal Structure Of The Far-Red Fluorescent Protein Hcred: Inherent Conformational Flexibility Of The Chromophore
Organism: Heteractis crispa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-05-17 Classification: LUMINESCENT PROTEIN Ligands: PEG |

