Search Count: 3,199
 |
Crystal Structure Of Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Pentaglutamate Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: EDO, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Bromodomain Containing Protein 1 With Crystal Epitope Mutations P566E:V569R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: GENE REGULATION |
 |
Jumonji Domain-Containing Protein 2B With Crystallization Epitope Mutations L916G:R917A:A918D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: EDO, SO4 |
 |
Protease From Norovirus Sydney Gii.4 Strain With Crystallization Epitope Mutation H50Y
Organism: Norovirus sydney 2212
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN |
 |
Jumonji Domain-Containing Protein 2B With Crystallization Epitope Mutations L916G:R917A:A918D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, NH4 |
 |
Jumonji Domain-Containing Protein 2D With Crystallization Epitope Mutation Q41R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: ZN, NI |
 |
Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Crystallization Epitope Mutations N221R:M226H
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL CYCLE Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: A1CY9, NA, UNX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: A1CU7, NA, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: PROTEIN TRANSPORT Ligands: A1CVQ, EDO |
 |
Spindlin Family Member 4 With Crystallization Epitope Mutations G129S:H131E
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: PROTEIN TRANSPORT Ligands: A1CSX, UNX |
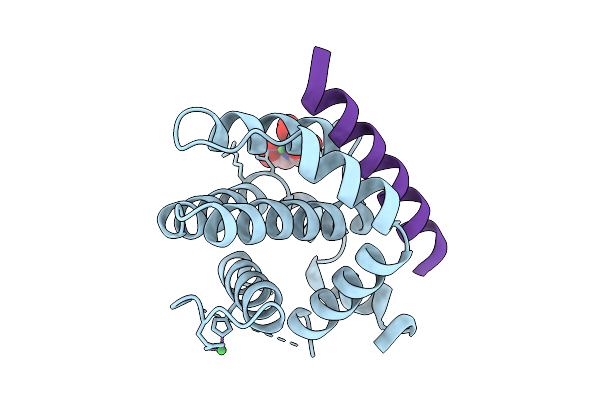 |
Crystal Structure Of Bcl-2 In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: APOPTOSIS Ligands: NI, NTA |
 |
Crystal Structure Of Bcl-2 (G101V) Mutant In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
|
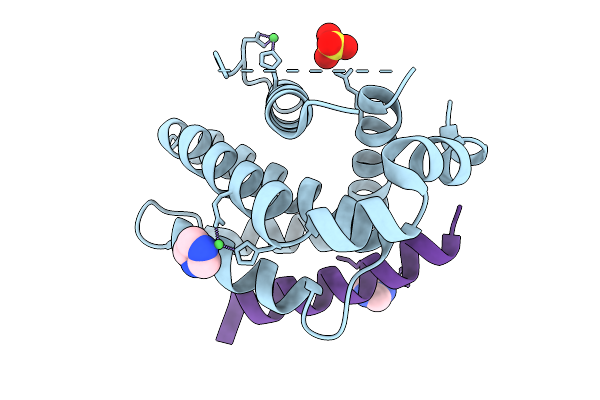 |
Crystal Structure Of Human Bcl-2 (R129L) Mutant In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: APOPTOSIS Ligands: IMD, SO4, NI |
 |
Organism: Mycolicibacterium
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE Ligands: GOL, MG, 8K6, SO4 |

