Search Count: 47
 |
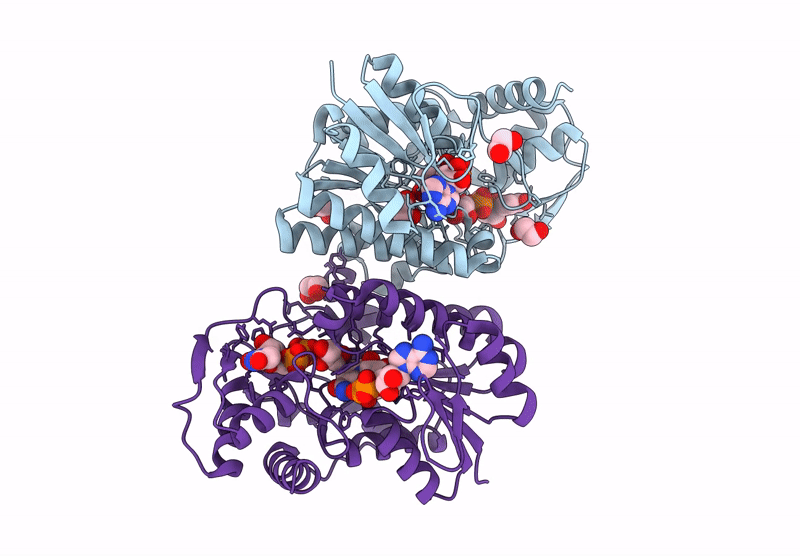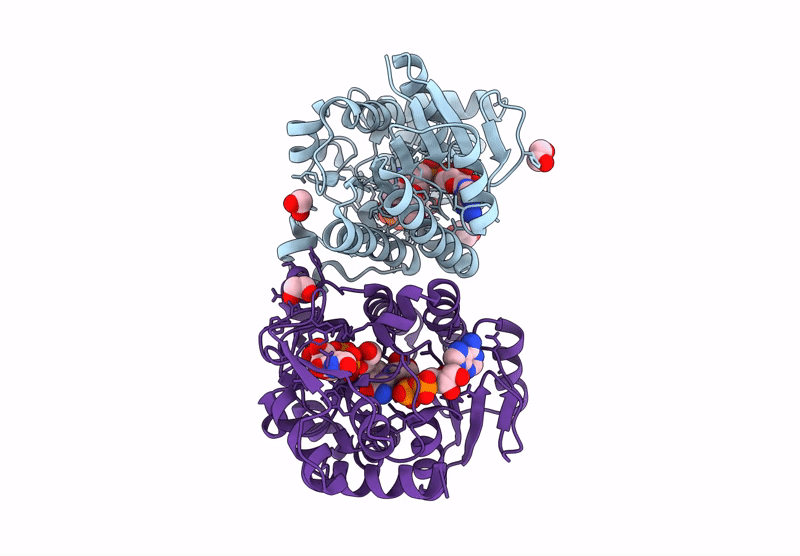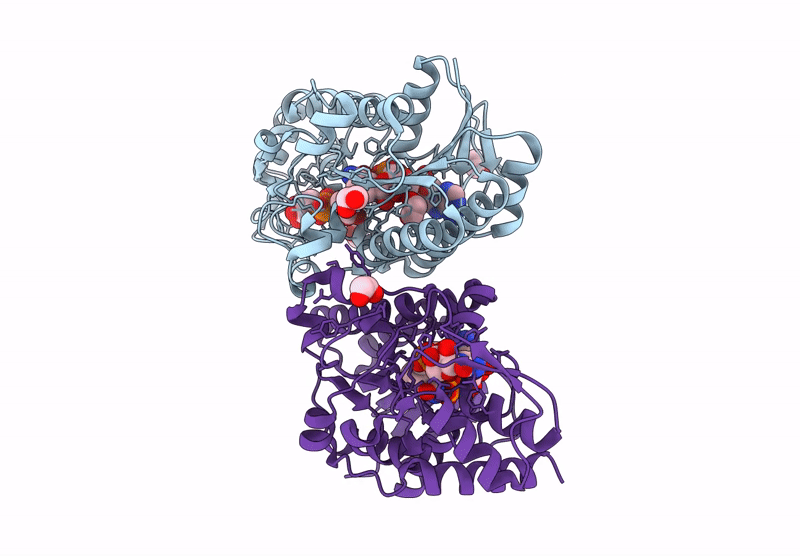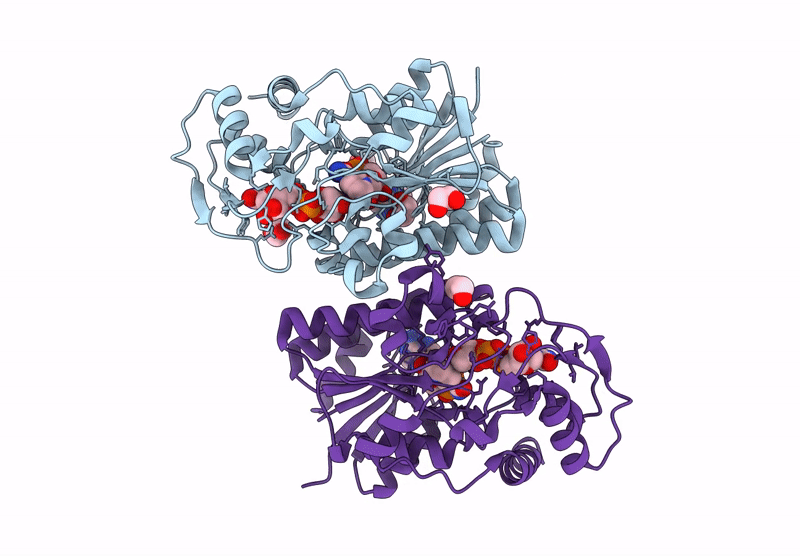
Structure Of Epimerase Mth373 From The Thermophilic Pseudomurein-Containing Methanogen Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-26 Classification: SUGAR BINDING PROTEIN Ligands: UDP, NAD, EDO, MG, CL |
 |
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-01-29 Classification: SUGAR BINDING PROTEIN Ligands: NAD, EDO, UDX, UDP, GOL |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Structure Of Mcrd (Methyl-Coenzyme M Reductase Operon Protein D) From Methanomassiliicoccus Luminyensis
Organism: Methanomassiliicoccus luminyensis b10
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-07-03 Classification: UNKNOWN FUNCTION Ligands: GOL |
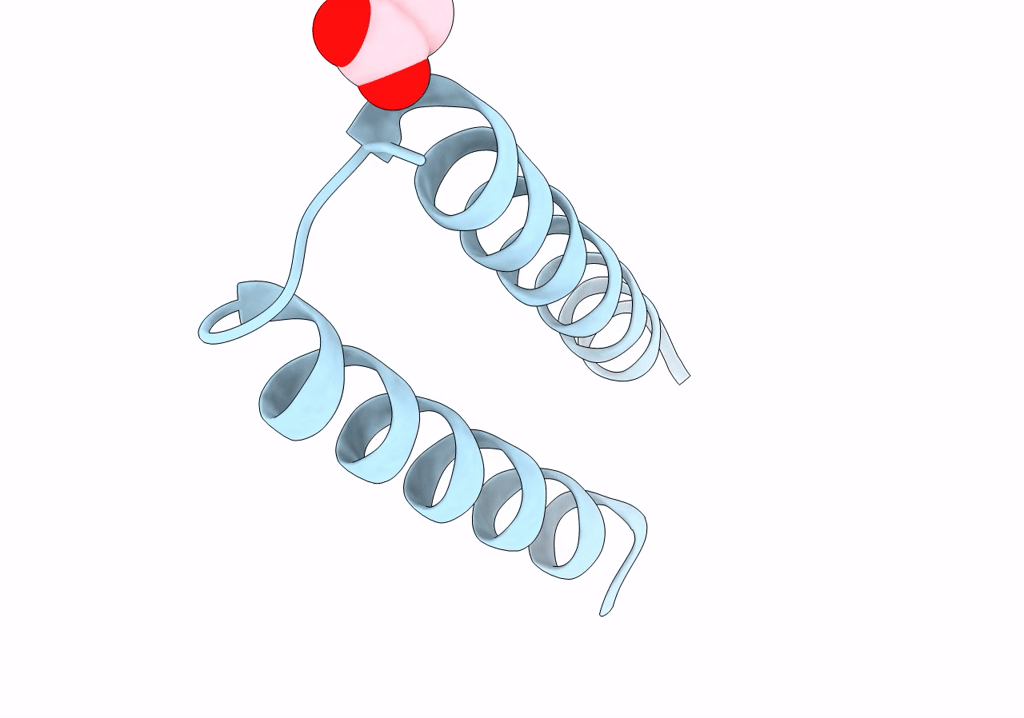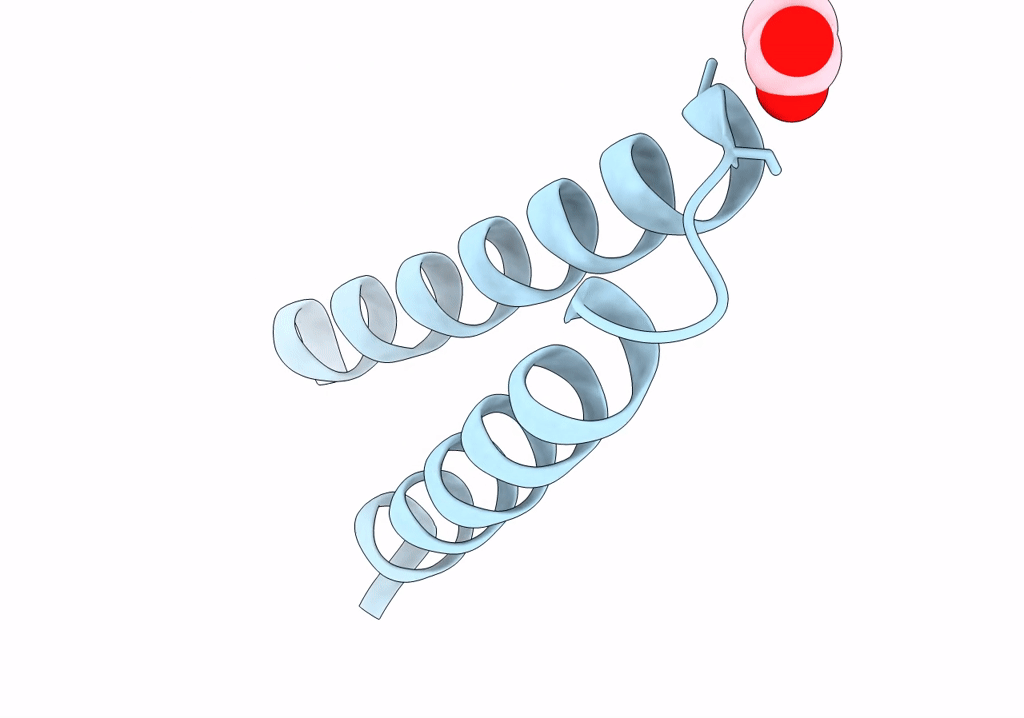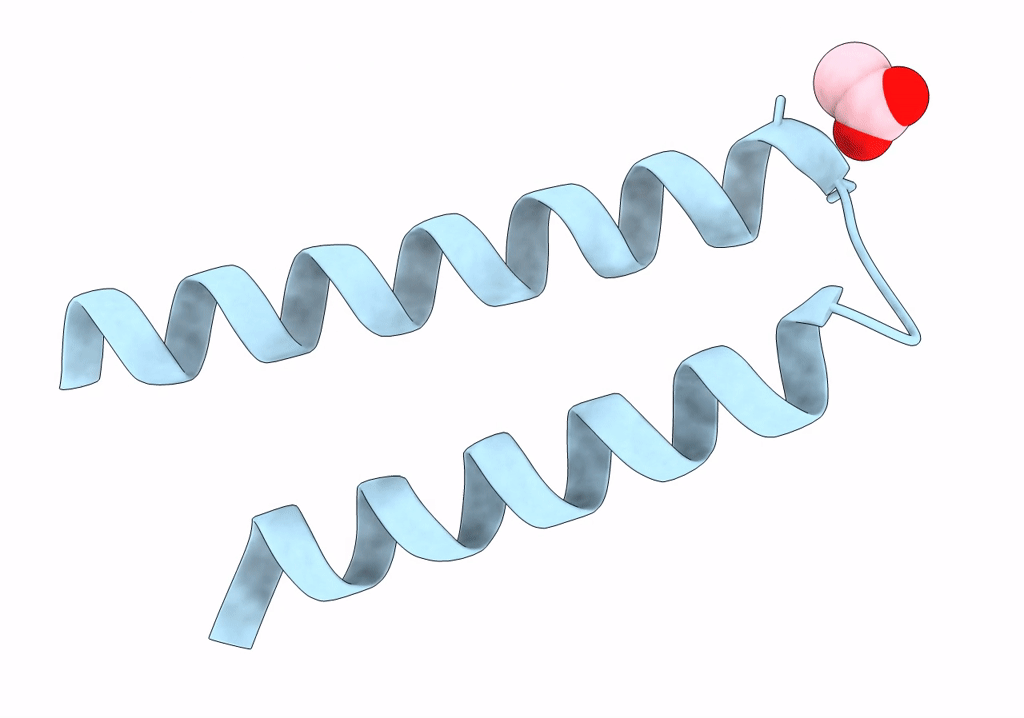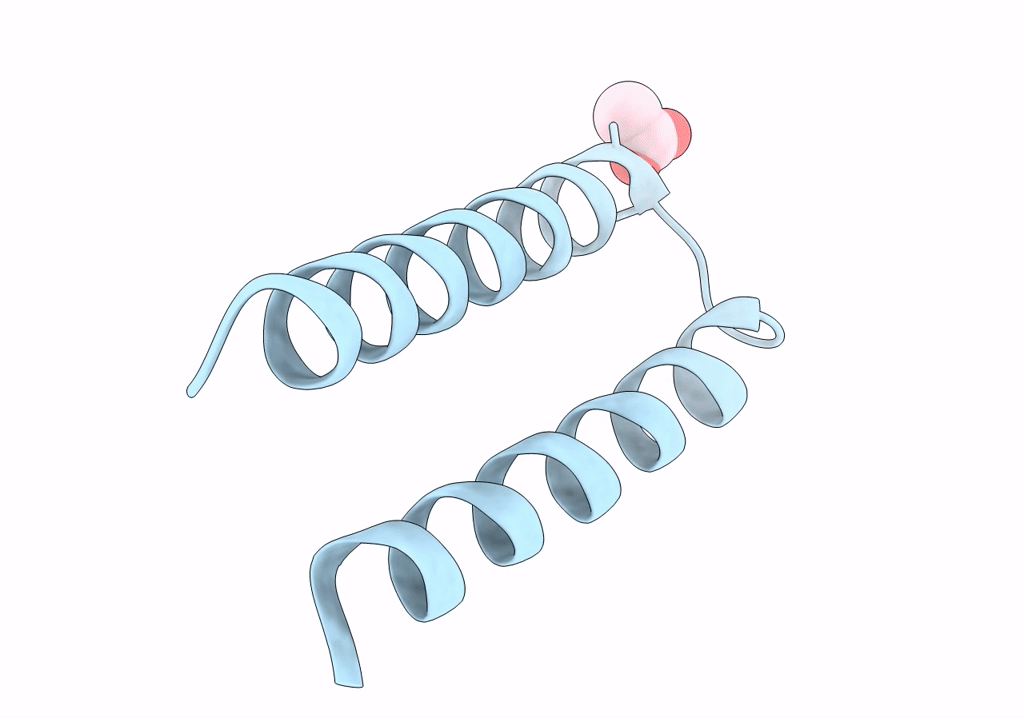 |
X-Ray Crystal Structure Of A De Novo Designed Helix-Loop-Helix Homodimer In An Anti Arrangement, Cc-Hp1.0
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-07 Classification: DE NOVO PROTEIN Ligands: ACT |
 |
X-Ray Crystal Structure Of A De Novo Selected Helix-Loop-Helix Heterodimer In A Syn Arrangement, 26Alpha/26Beta
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: DE NOVO PROTEIN Ligands: SO4, GOL, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: UNL, MES, NAG |
 |
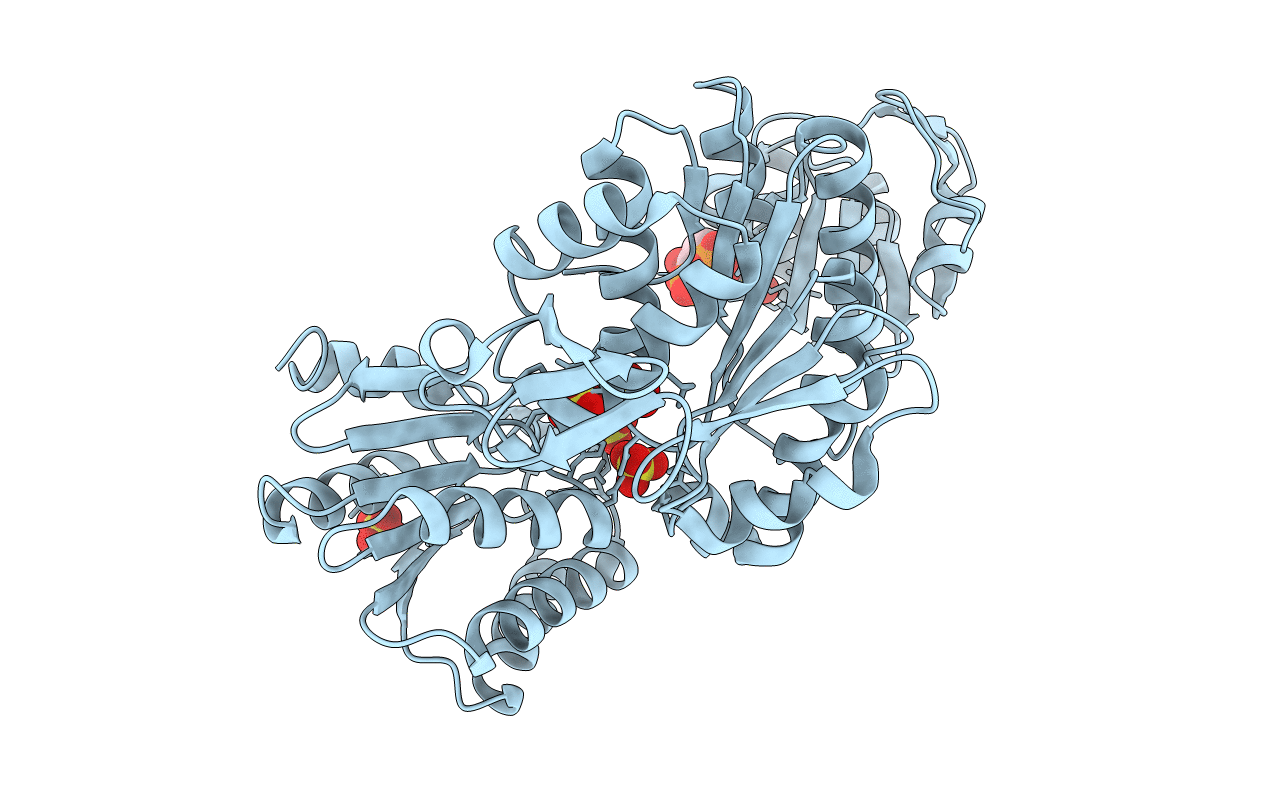
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2022-10-12 Classification: LIGASE |
 |
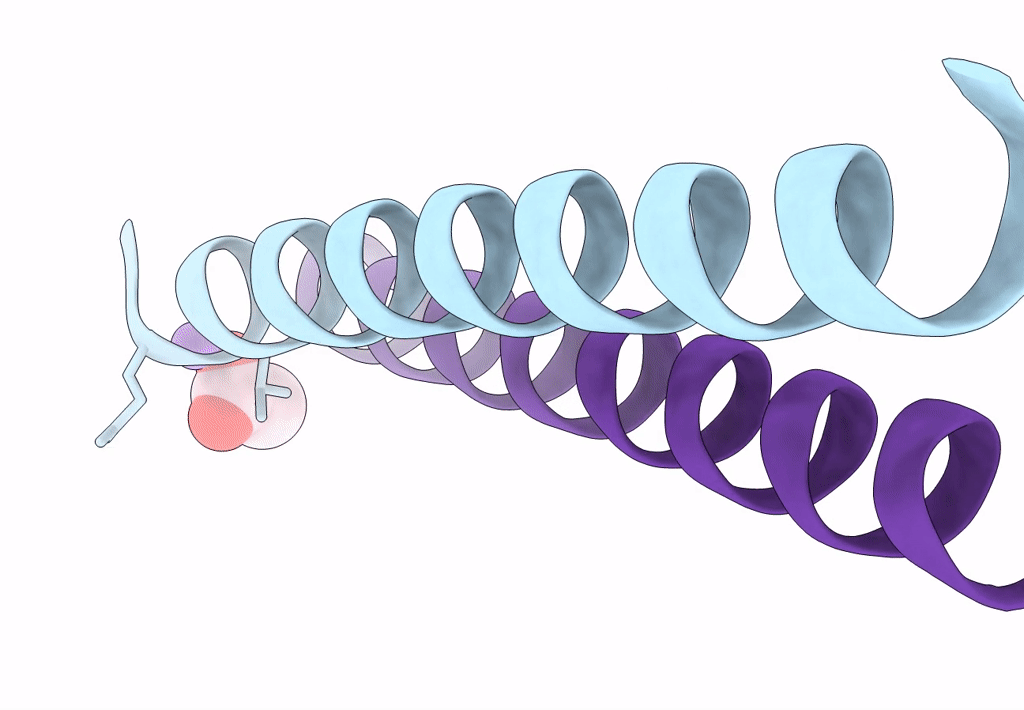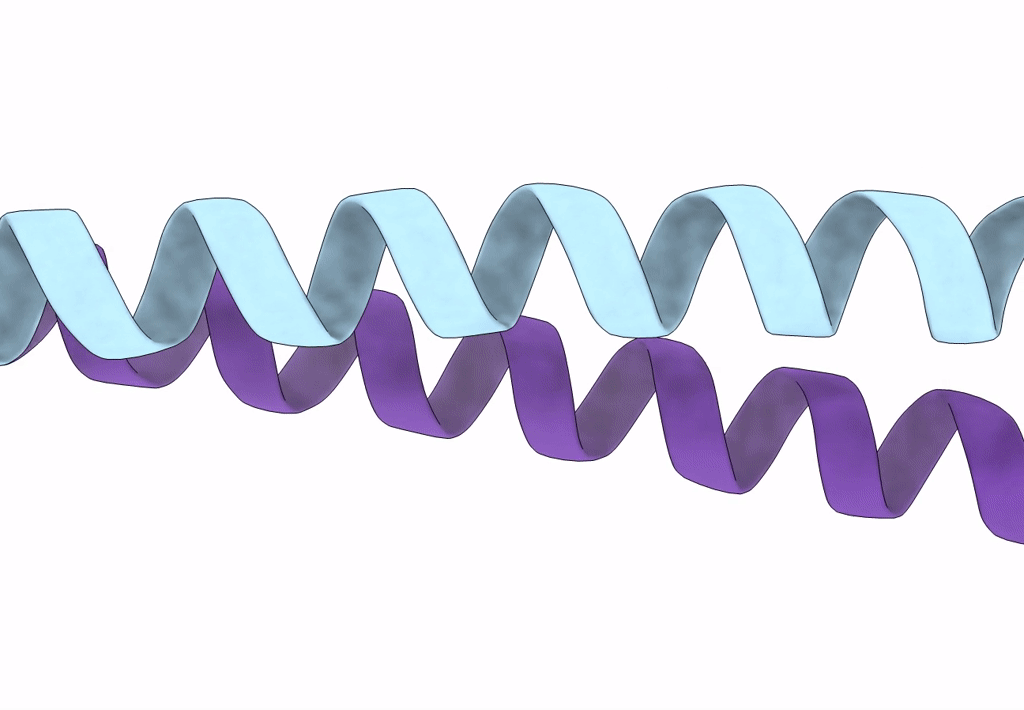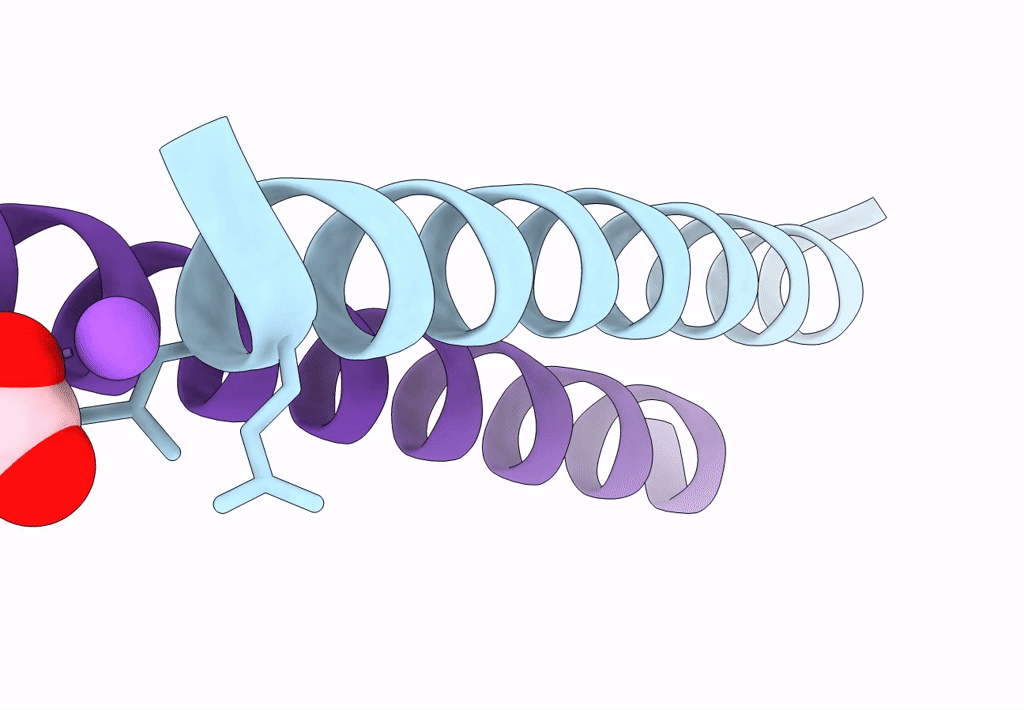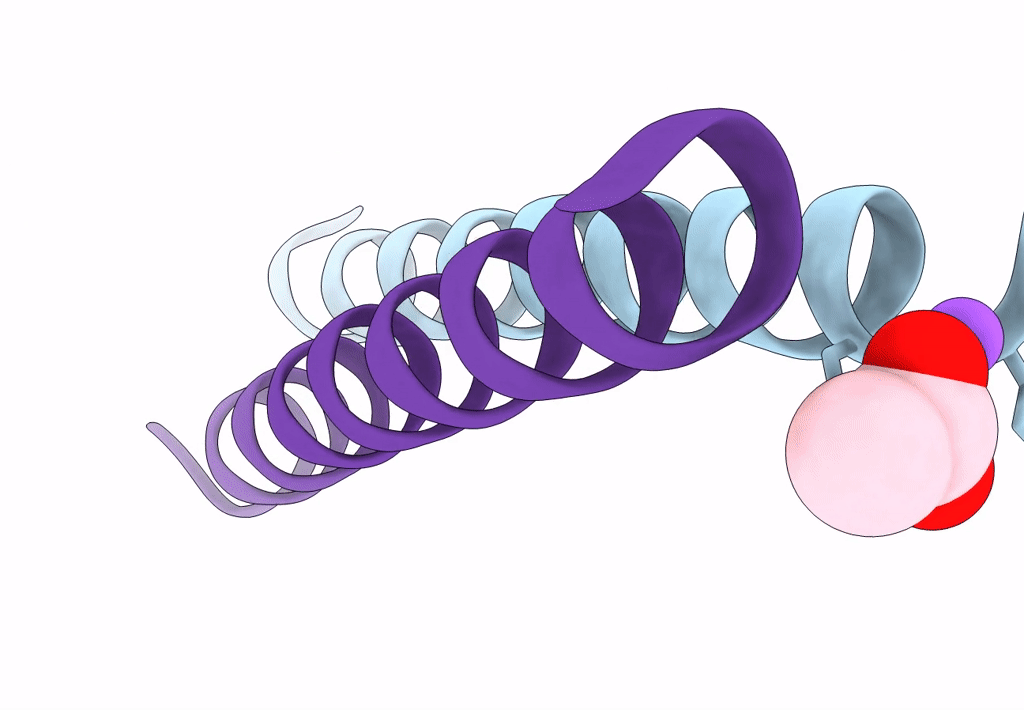
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-12 Classification: LIGASE Ligands: UDP, SO4 |
 |
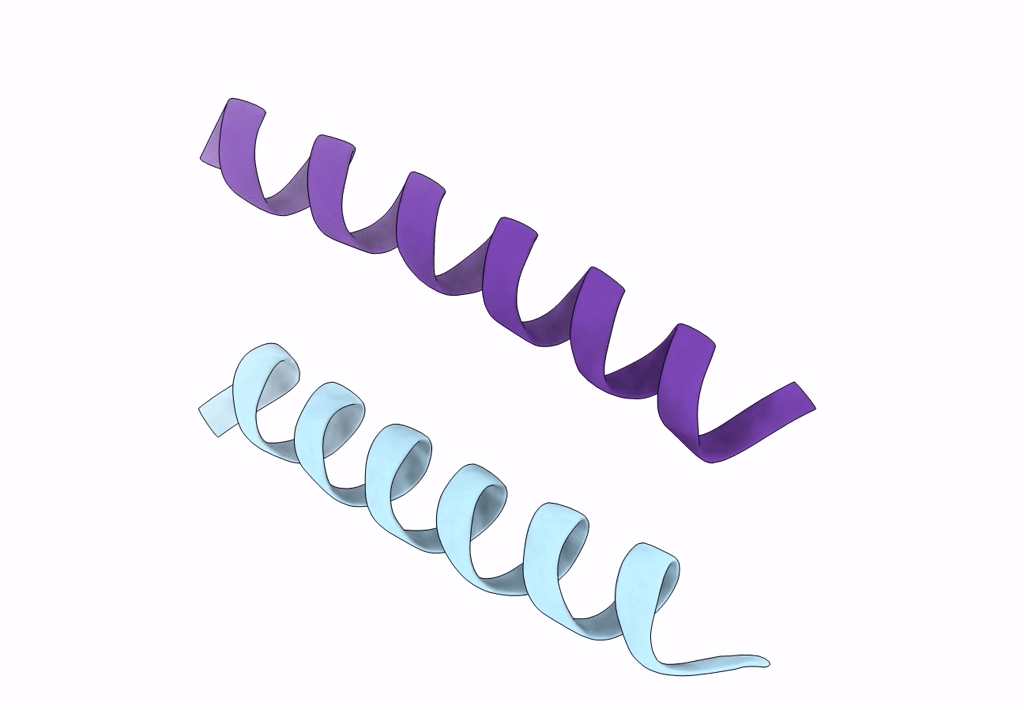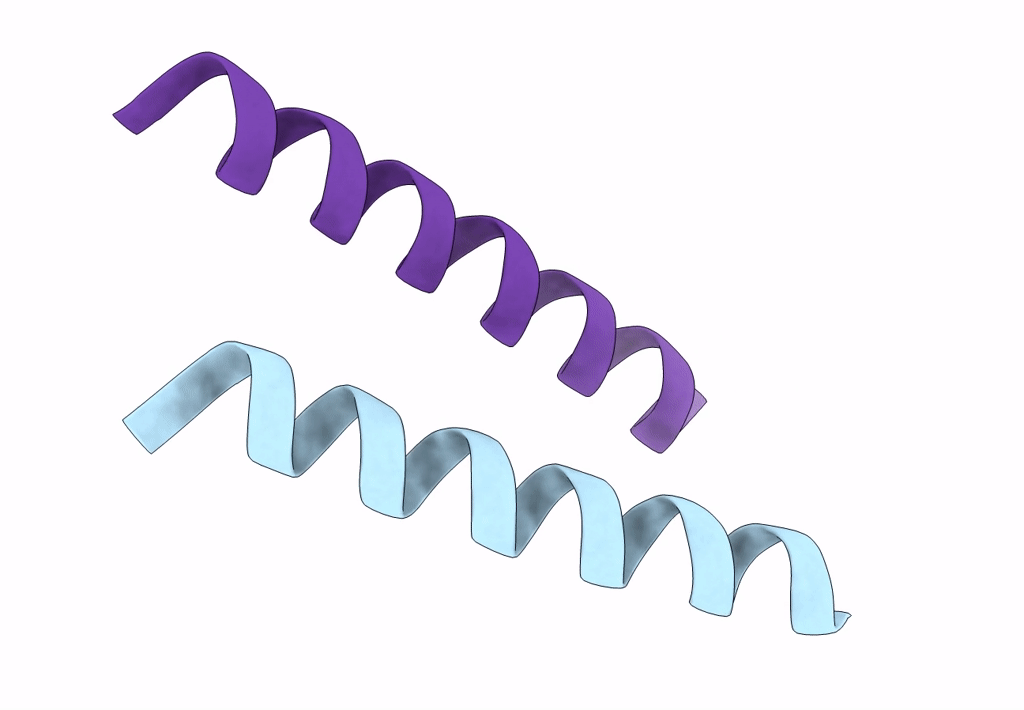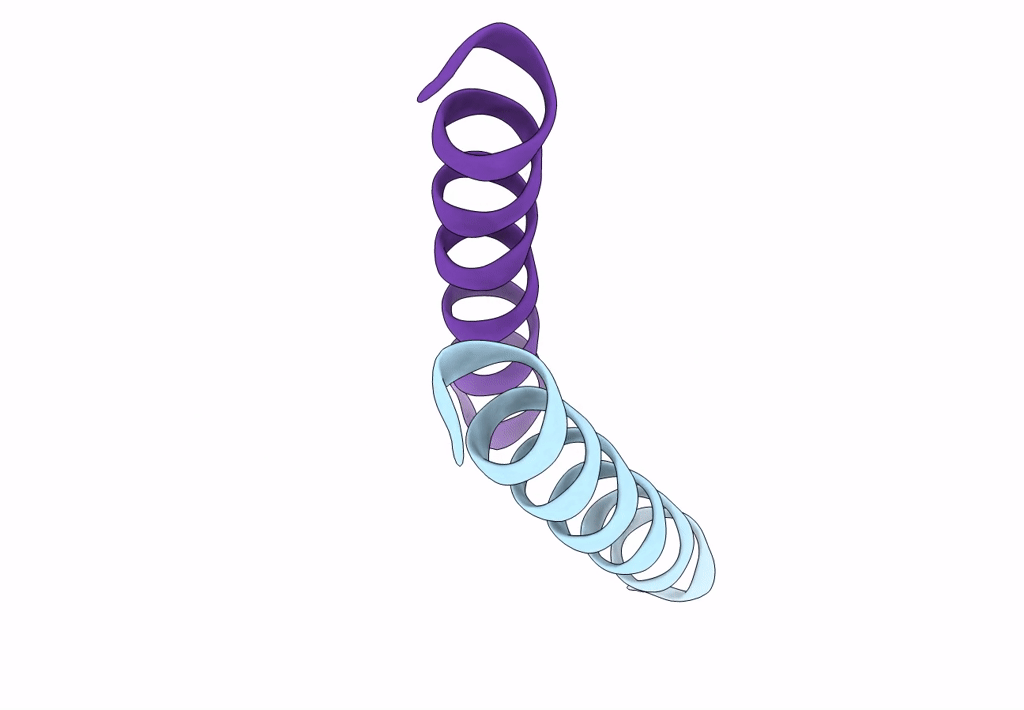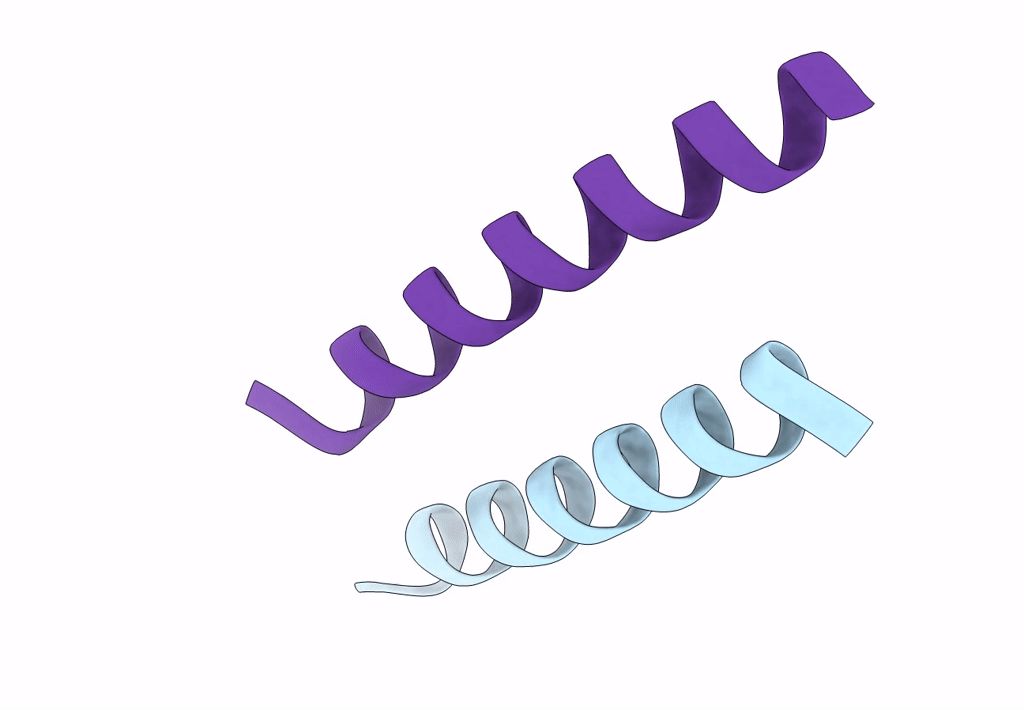
X-Ray Crystal Structure Of A De Novo Designed Antiparallel Coiled-Coil Homotetramer With 4 Heptad Repeats, Apcc-Tet*
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2022-10-05 Classification: DE NOVO PROTEIN Ligands: ACT, NA |
 |
X-Ray Crystal Structure Of A De Novo Designed Antiparallel Coiled-Coil Homotetramer With 3 Heptad Repeats, Apcc-Tet*3
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-10-05 Classification: DE NOVO PROTEIN |
 |
X-Ray Crystal Structure Of A De Novo Designed Antiparallel Coiled-Coil Heterotetramer With 3 Heptad Repeats, Apcc-Tet*3-A2B2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-05 Classification: DE NOVO PROTEIN |
 |
X-Ray Crystal Structure Of A De Novo Designed Single-Chain Antiparallel 4-Helix Coiled-Coil Bundle, Sc-Apcc-4
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-05 Classification: DE NOVO PROTEIN |
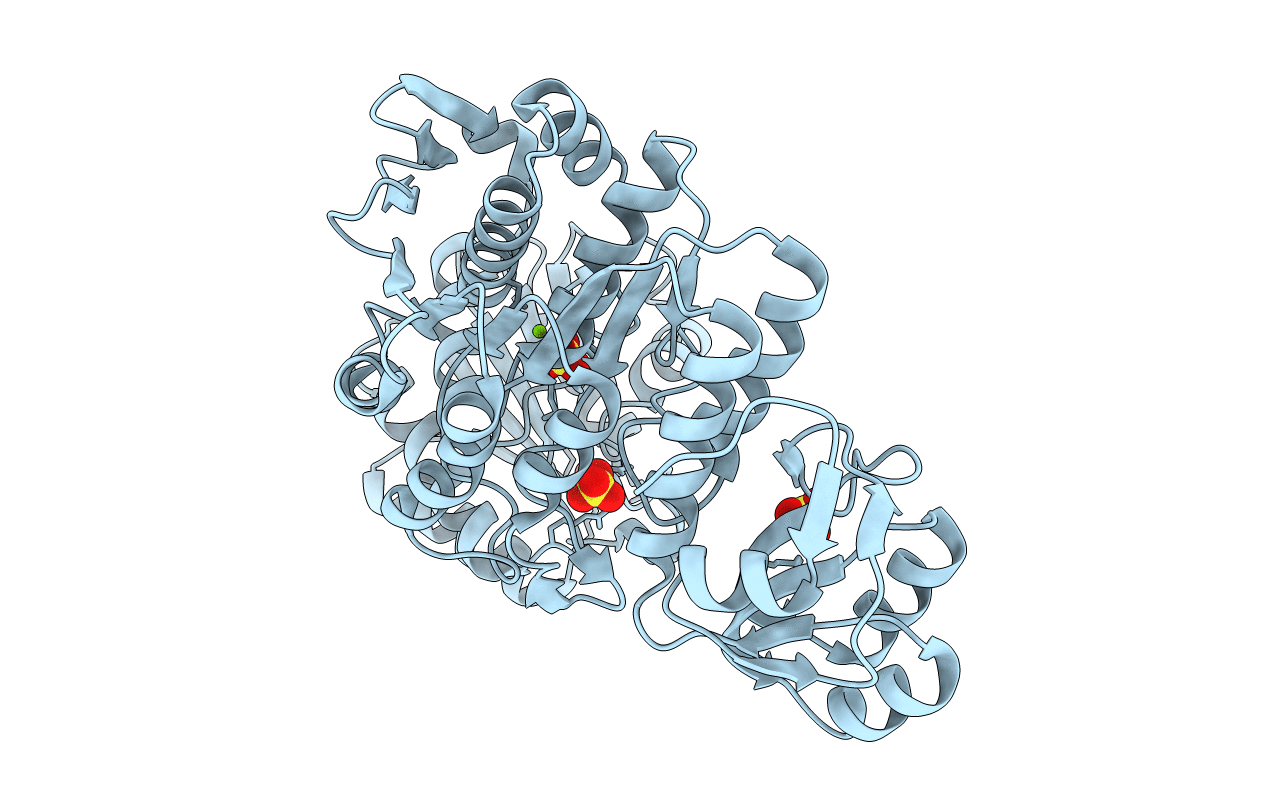 |
Apo Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus (strain atcc 43054 / dsm 2088 / jcm 10308 / v24 s)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-09-01 Classification: LIGASE Ligands: MG, SO4 |
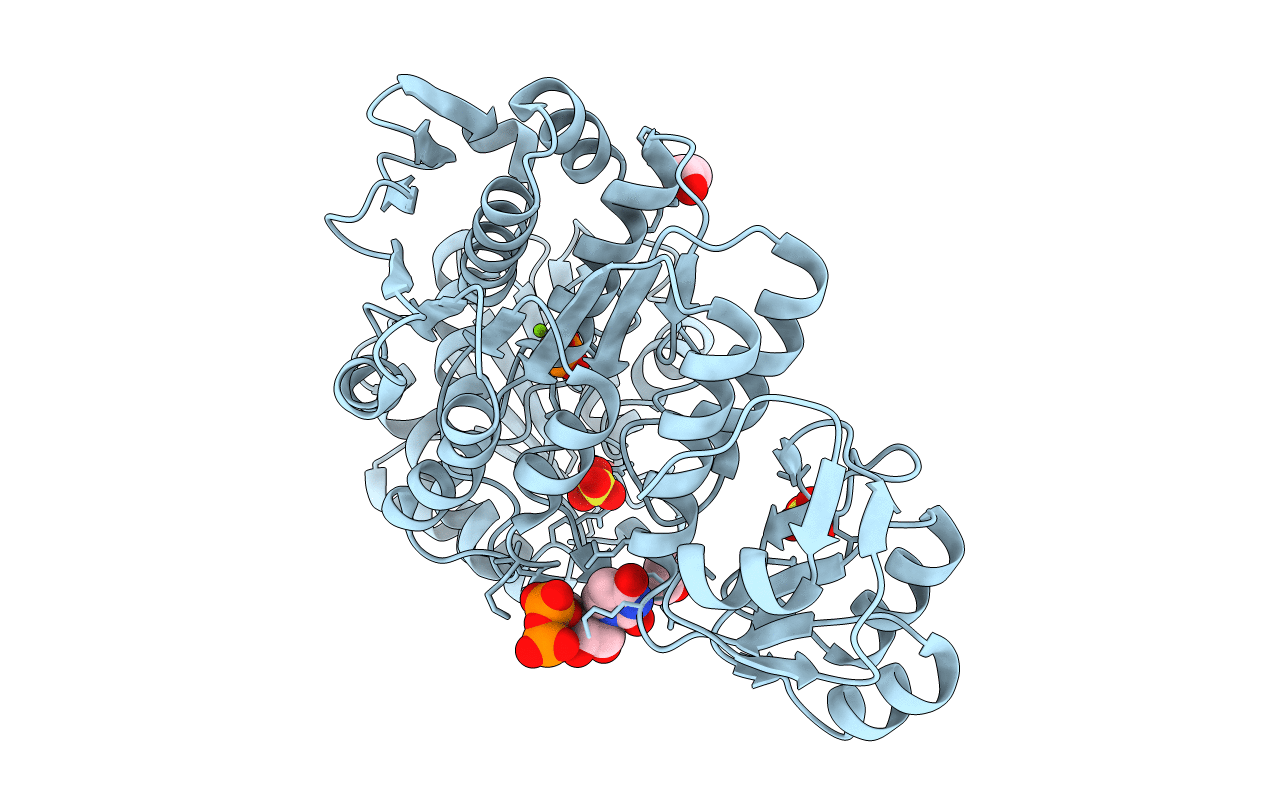 |
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: LIGASE Ligands: PO4, MG, EDO, SO4, UDP |
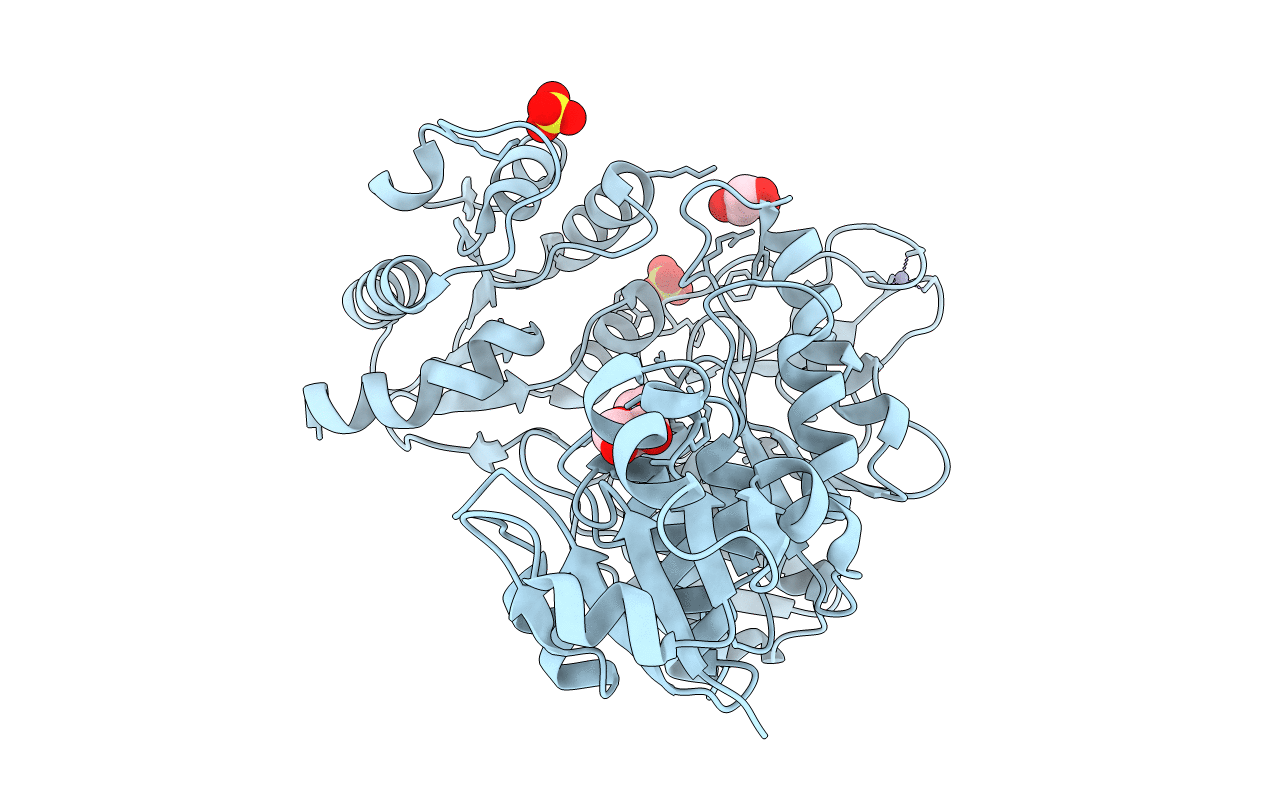 |
Structure Of A Pseudomurein Peptide Ligase Type C From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-10 Classification: LIGASE Ligands: SO4, ZN, GOL, AAE |
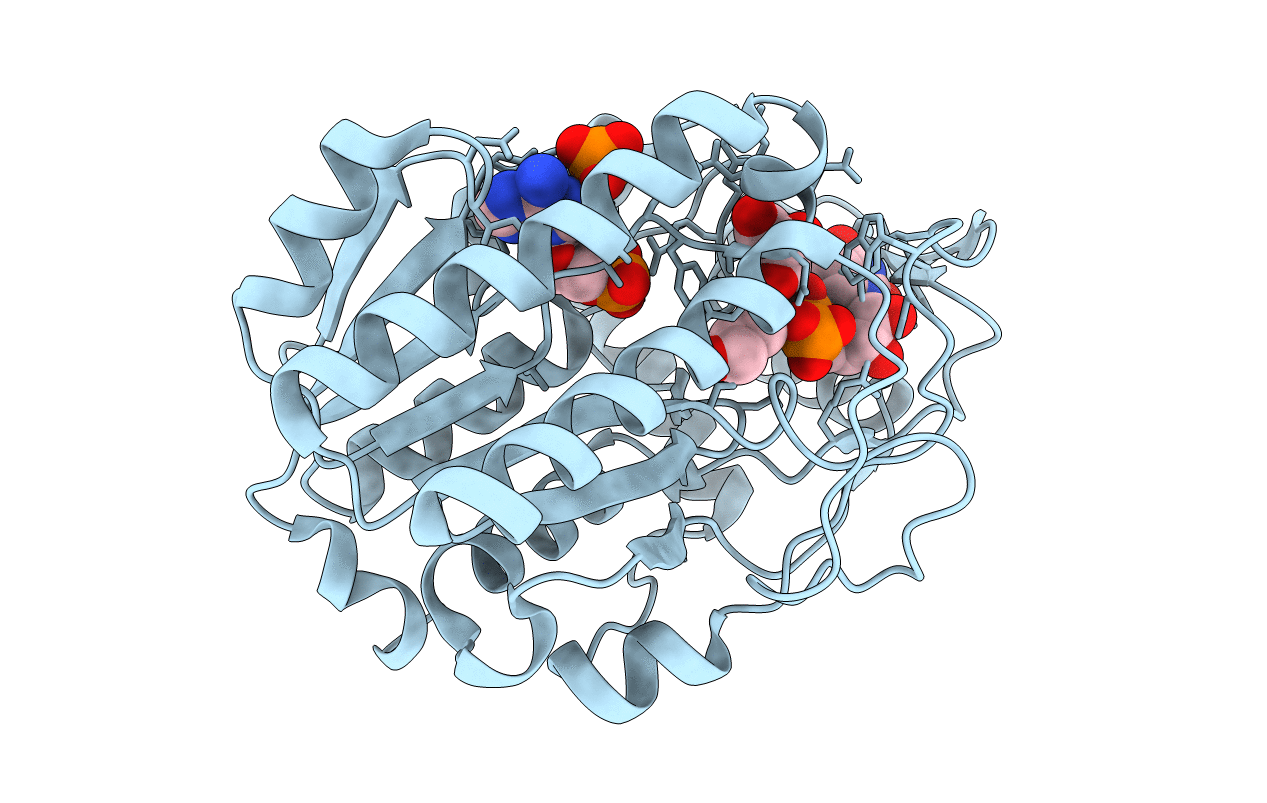 |
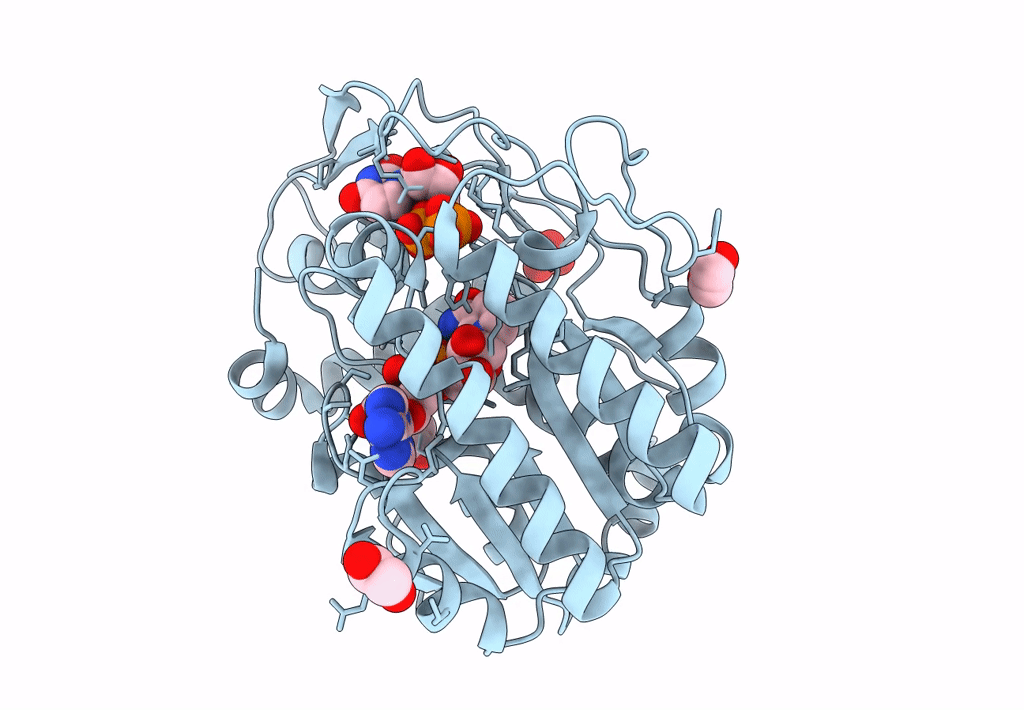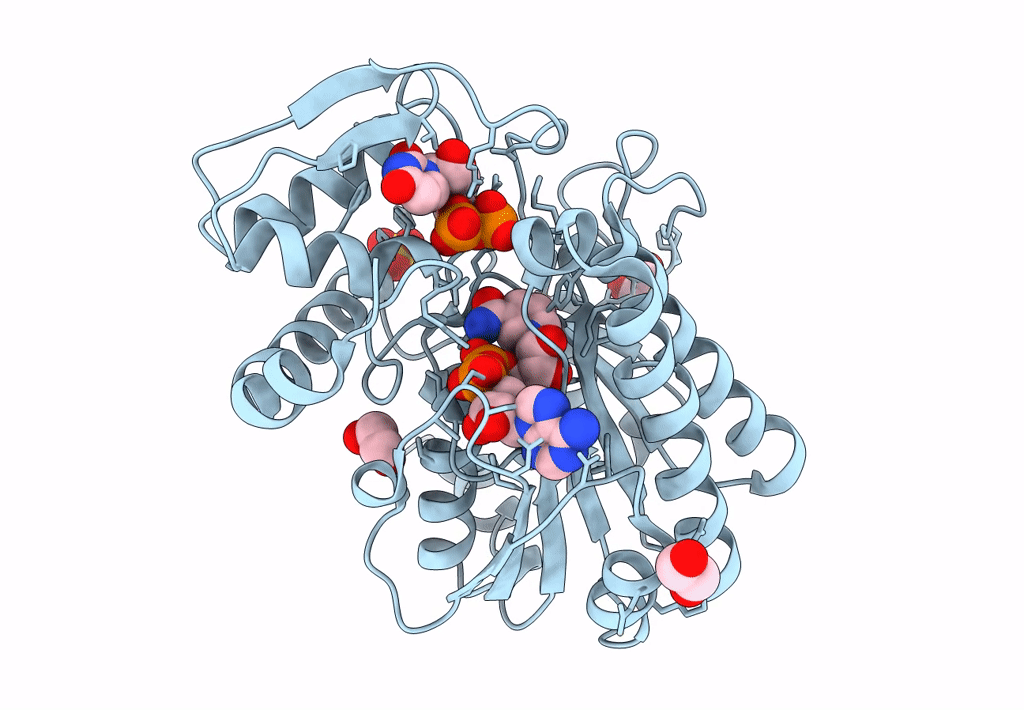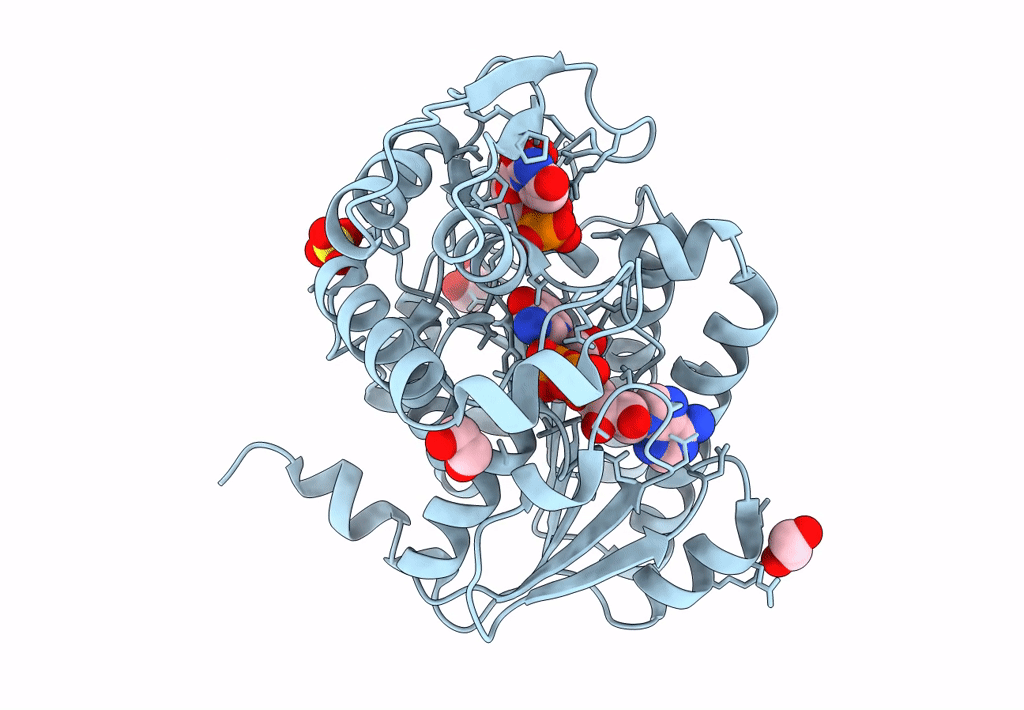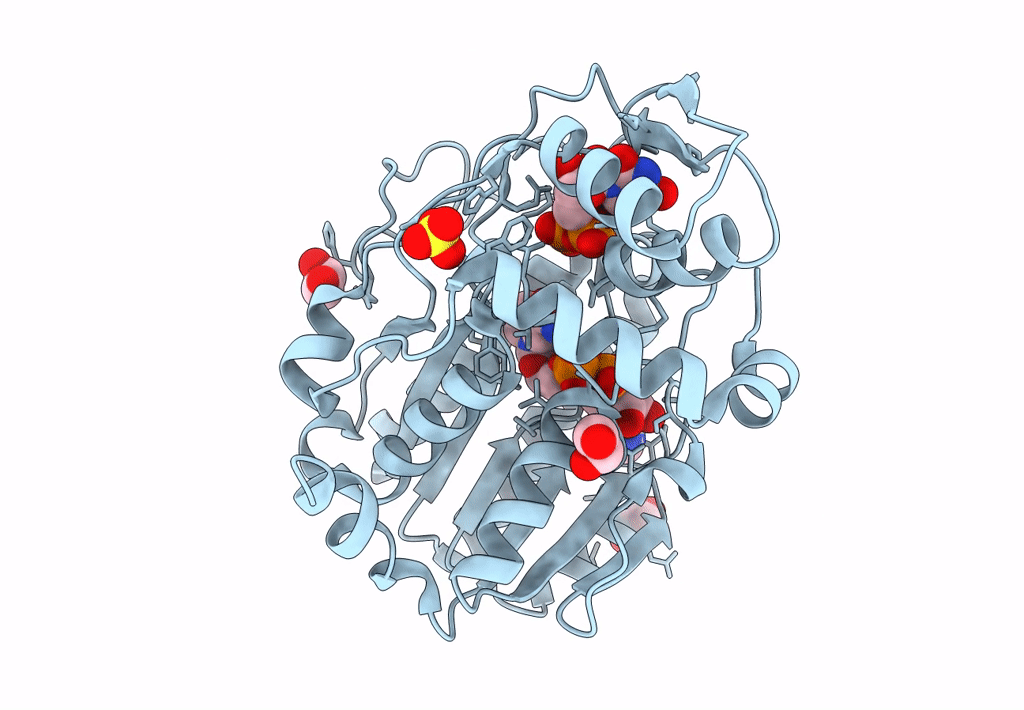
Structure Of Epimerase Mth375 From The Thermophilic Pseudomurein-Containing Methanogen Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-07-08 Classification: ISOMERASE Ligands: PO4, GOL, CL, UDP, AMP |
 |
Structure Of Epimerase Mth375 From The Thermophilic Pseudomurein-Containing Methanogen Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-08 Classification: ISOMERASE Ligands: NAD, UDP, EDO, SO4 |
 |
2,5-Diamino-6-(Ribosylamino)-4(3H)-Pyrimidinone 5'-Phosphate Reductase (Mthred) From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: CL, GOL, MG, NAP |

