Search Count: 19
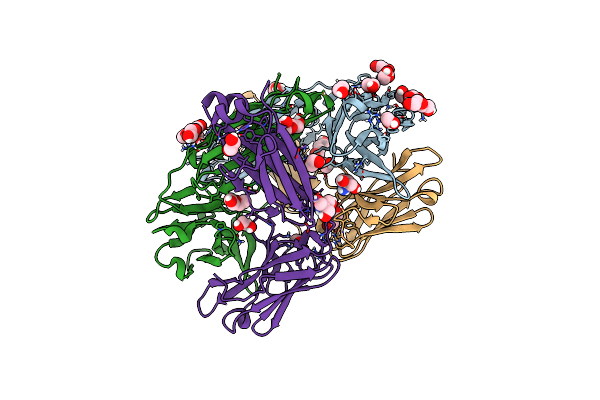 |
Structure Of The Arginase-2-Inhibitory Human Antigen-Binding Fragment Fab C0021144
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: CL, EDO, PEG, TRS |
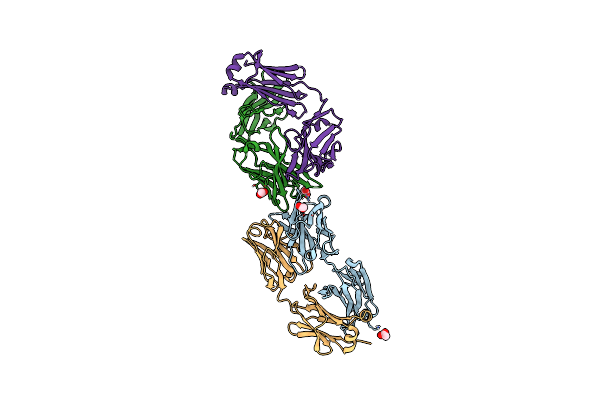 |
Structure Of The Arginase-2-Inhibitory Human Antigen-Binding Fragment Fab C0021158
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: ACT, CL |
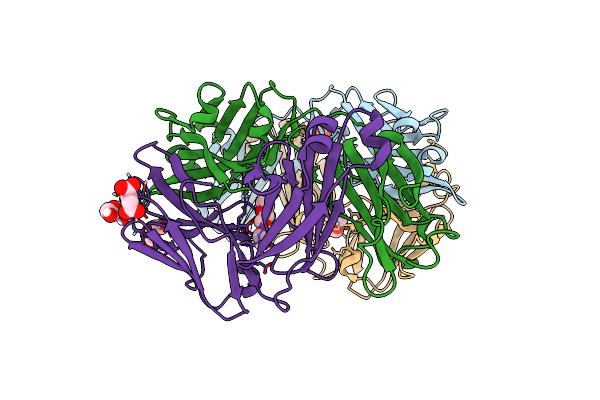 |
Structure Of The Arginase-2-Inhibitory Human Antigen-Binding Fragment Fab C0021181
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: CL, SIN, PEG, PGE |
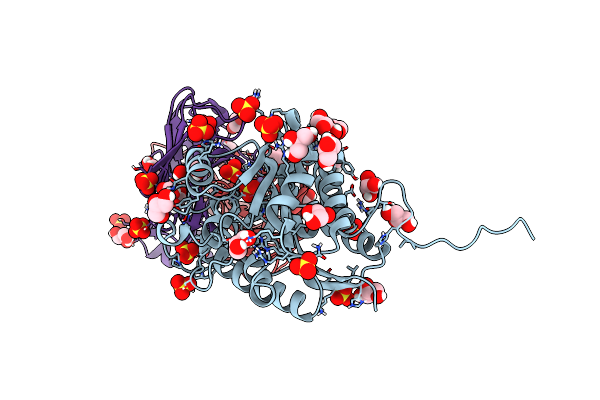 |
Structure Of Arginase-2 In Complex With The Inhibitory Human Antigen-Binding Fragment Fab C0021158
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: GOL, MN, SO4 |
 |
Structure Of Arginase-2 In Complex With The Inhibitory Human Antigen-Binding Fragment Fab C0021181
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: MN, PO4 |
 |
Structure Of The Arginase-2-Inhibitory Human Antigen-Binding Fragment Fab C0020187
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: CL, MPD, EPE, SO4 |
 |
Structure Of Arginase-2 In Complex With The Inhibitory Human Antigen-Binding Fragment Fab C0020187
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: MN, SO4 |
 |
Structure Of The Arginase-2-Inhibitory Human Antigen-Binding Fragment Fab C0021177
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-06-10 Classification: PROTEIN BINDING Ligands: PG4, P6G, MLT, PGE |
 |
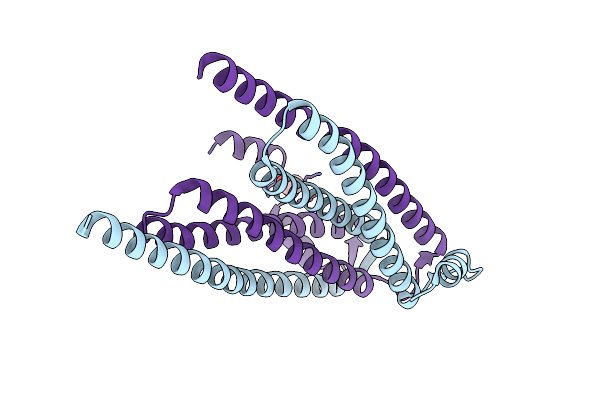
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2017-04-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PG4 |
 |
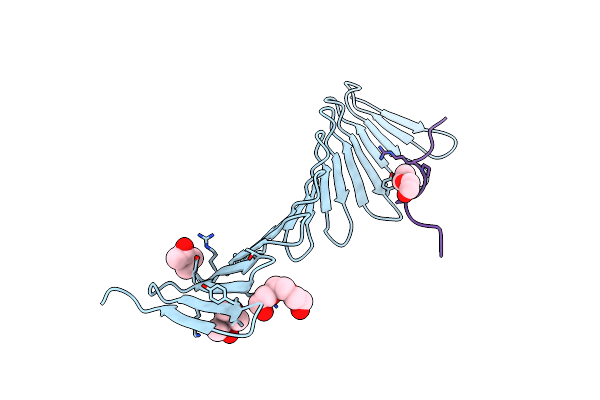
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-04-16 Classification: PROTEIN BINDING Ligands: CL, ACT |
 |
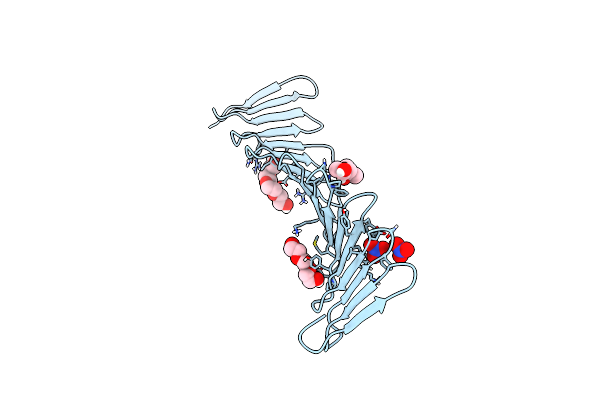
A Novel Domain In The Microcephaly Protein Cpap Suggests A Role In Centriole Architecture
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-09-11 Classification: structural protein, protein binding Ligands: PGE, MPD, PG4 |
 |
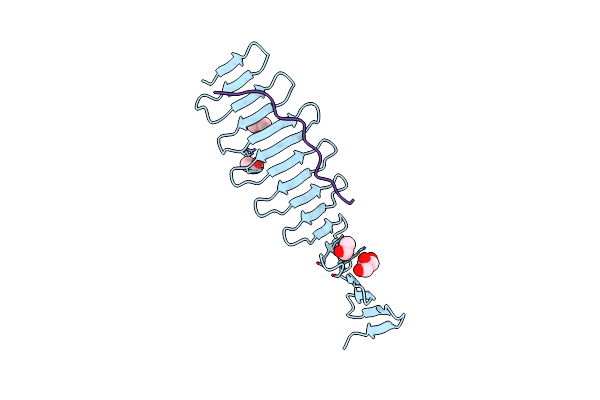
Structural Analysis Of The Microcephaly Protein Cpap G-Box Domain Suggests A Role In Centriole Elongation.
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2013-09-04 Classification: structural protein, protein binding Ligands: NO3, PGE, PG4, MPD |
 |
Structural Analysis Of The Microcephaly Protein Cpap G-Box Domain Suggests A Role In Centriole Elongation.
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2013-09-04 Classification: structural protein, protein binding Ligands: IPA, GOL |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-06-19 Classification: STRUCTURAL PROTEIN Ligands: GOL, PG4 |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-06-19 Classification: STRUCTURAL PROTEIN Ligands: GOL, PG4 |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2013-06-19 Classification: STRUCTURAL PROTEIN Ligands: MPD |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2013-06-19 Classification: STRUCTURAL PROTEIN Ligands: EDO, IMD, PG4 |
 |
Organism: Plasmodium falciparum
Method: SOLUTION NMR Release Date: 2012-01-25 Classification: CELL ADHESION |
 |
Organism: Schizosaccharomyces pombe
Method: SOLUTION NMR Release Date: 2003-07-10 Classification: NUCLEAR PROTEIN |

