Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
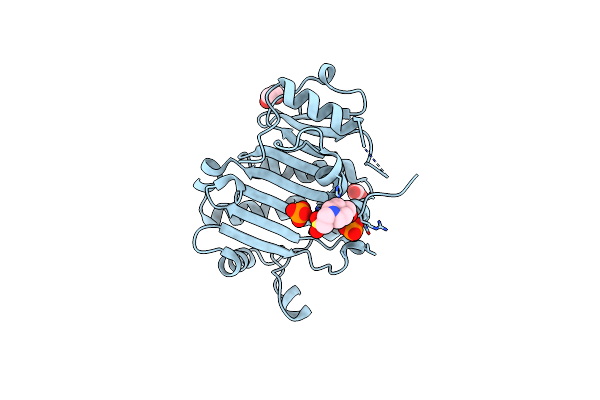 |
Crystal Structure Of The Immunity Protein (52 Domain-Containing Protein) From Pseudomonas Aeruginosa Pao1.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: ANTITOXIN Ligands: EPE, PO4, EDO, CO3 |
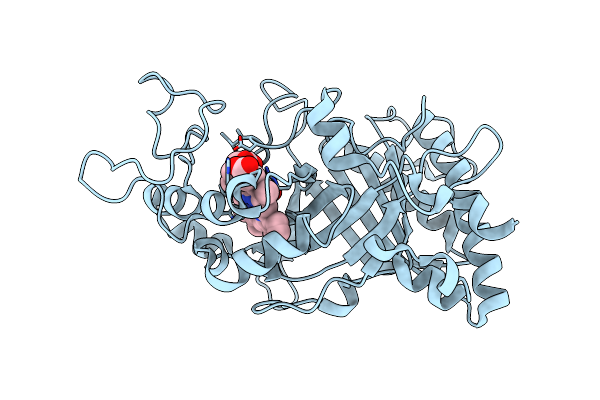 |
Organism: Frankia casuarinae
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, OXY |
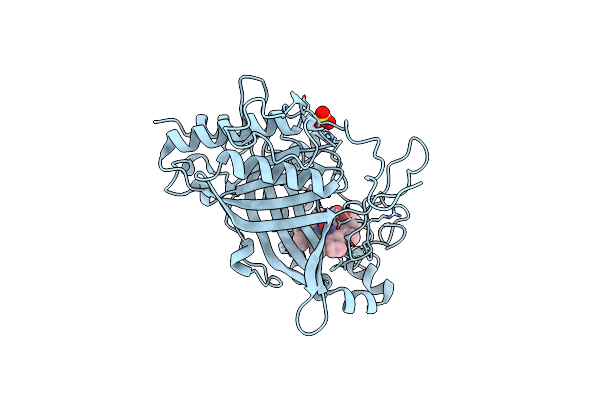 |
Organism: Pseudomonas rhizosphaerae
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, CL, SO4 |
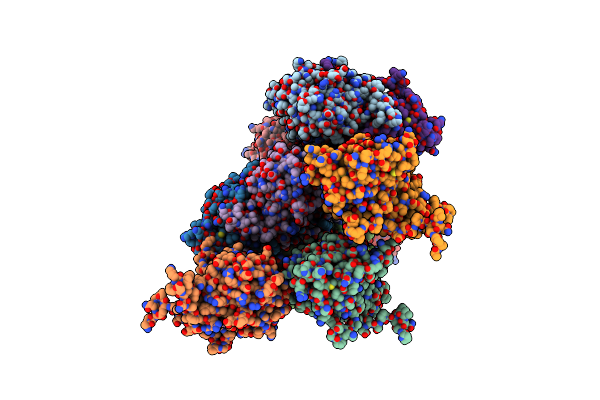 |
Organism: Pseudomonas fluorescens (strain atcc baa-477 / nrrl b-23932 / pf-5)
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, OXY |
 |
Crystal Structure Of Cib_12 Beta-Galactosidase From Cuniculiplasma Divulgatum
Organism: Cuniculiplasma divulgatum
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Cib_13 Beta-Galactosidase From Cuniculiplasma Divulgatum
Organism: Cuniculiplasma divulgatum
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: GOL, TRS |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
 |
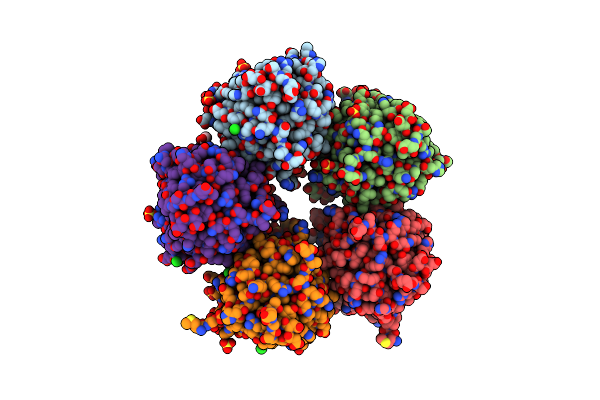
Crystal Structure Of Fluoroacetate Dehalogenase Daro3835 H274N Mutant With D107-Glycolyl Intermediate
Organism: Dechloromonas aromatica rcb
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-14 Classification: HYDROLASE |
 |
Organism: Dechloromonas aromatica rcb
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-09-06 Classification: HYDROLASE Ligands: CL |
 |
Receptor Shhtl5 From Striga Hermonthica In Complex With Strigolactone Agonist Gr24
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2023-06-14 Classification: SIGNALING PROTEIN Ligands: TZU, SO4, EDO, CL |
 |
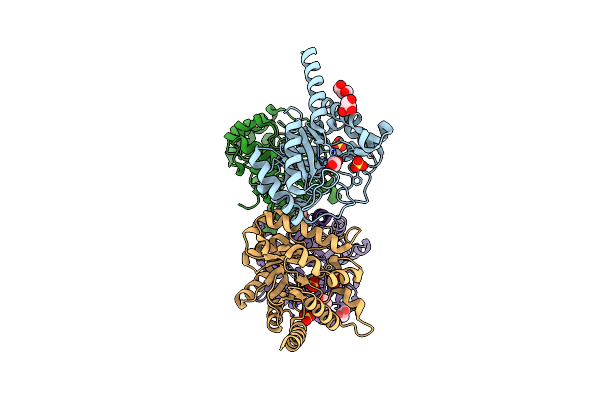
Crystal Structure Of Sulfonamide Resistance Enzyme Sul1 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR, GOL, SO4, CL |
 |
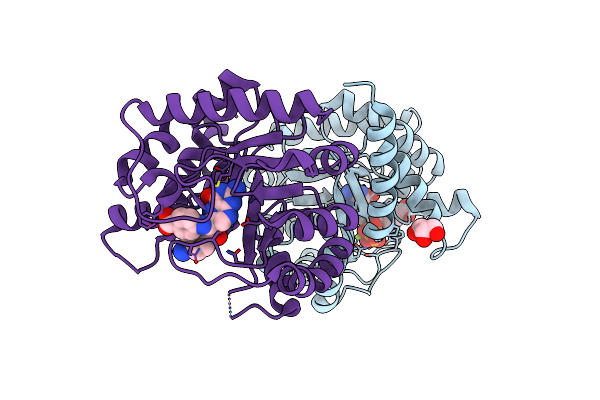
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: SO4, CL, GOL, PEG |
 |
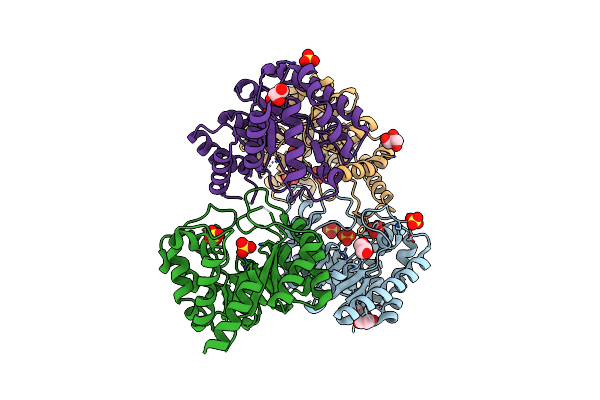
Crystal Structure Of Sulfonamide Resistance Enzyme Sul2 In Complex With 7,8-Dihydropteroate, Magnesium, And Pyrophosphate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: 78H, POP, MG, GOL, CL, PAB |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: GOL, CL, SO4 |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
Crystal Structure Of Adaptive Laboratory Evolved Sulfonamide-Resistant Dihydropteroate Synthase (Dhps) From Escherichia Coli In Complex With 6-Hydroxymethylpterin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With Reaction Intermediate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: XHP, PAB, GOL, CL, MG, POP |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-08-10 Classification: TRANSFERASE Ligands: EDO, FMT, OO9, SO4 |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: OXIDOREDUCTASE Ligands: FMN, CIT, PGE, EDO, PEG |