Search Count: 181
 |
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: CELL CYCLE Ligands: GTP, GDP, ADP, MG, ALF |
 |
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: CELL CYCLE Ligands: GTP, GDP, MG, ADP |
 |
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: CELL CYCLE Ligands: GTP, GDP, MG, ADP |
 |
Organism: Drosophila melanogaster, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: CELL CYCLE Ligands: GTP, GDP, ADP, MG, ALF |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: PLANT PROTEIN Ligands: ADP |
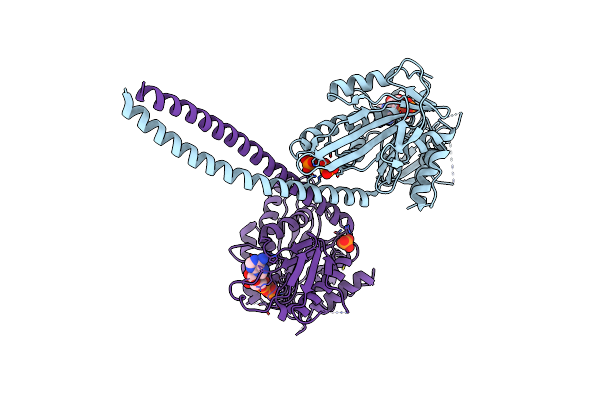 |
Crystal Structure Of The Kinesin-14 Motor Protein From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2025-04-09 Classification: MOTOR PROTEIN Ligands: ADP, PO4 |
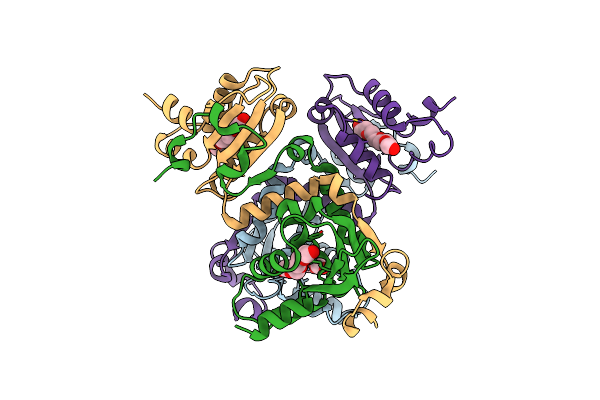 |
Structural Basis For The Distinct Roles Of Non-Conserved Pro116 And Conserved Tyr124 Of Bch Domain Of Yeast P50Rhogap
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-05-29 Classification: LIPID BINDING PROTEIN Ligands: PG4 |
 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: FE2, HEM |
 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: METAL BINDING PROTEIN Ligands: FE2, FE, HEM |
 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: METAL BINDING PROTEIN Ligands: FE2, FE, HEM |
 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: METAL BINDING PROTEIN Ligands: FE2, FE, HEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: ANTITUMOR PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: ANTITUMOR PROTEIN |
 |
Organism: Lachnospiraceae bacterium ma2020, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.95 Å Release Date: 2023-02-22 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Lachnospiraceae bacterium
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-02-08 Classification: DNA BINDING PROTEIN Ligands: SO4, MG |
 |
Near Full Length Kidney Type Glutaminase In Complex With 2,2-Dimethyl-2,3-Dihydrobenzo[A] Phenanthridin-4(1H)-One (Ddp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-12-21 Classification: ONCOPROTEIN Ligands: HZO |
 |
Organism: Chromobacterium haemolyticum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-07-20 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2022-07-06 Classification: IMMUNE SYSTEM Ligands: GOL, SO4 |
 |
Crystal Structure Of Human Microplasmin In Complex With Kazal-Type Inhibitor Aati
Organism: Aedes aegypti, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: GOL, NA |

