Search Count: 209
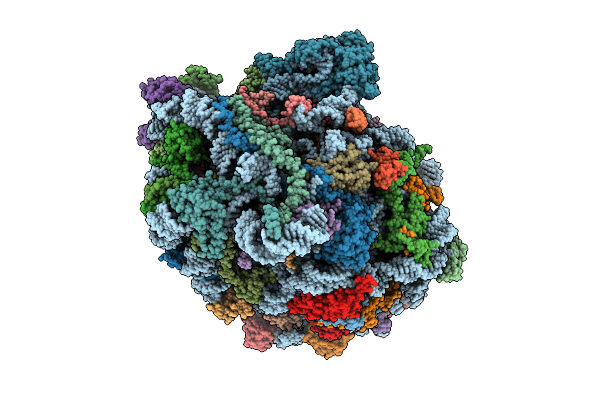 |
High Resolution Structure Of The Thermophilic 60S Ribosomal Subunit Of Chaetomium Thermophilum
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, SPM, MG, K, ZN |
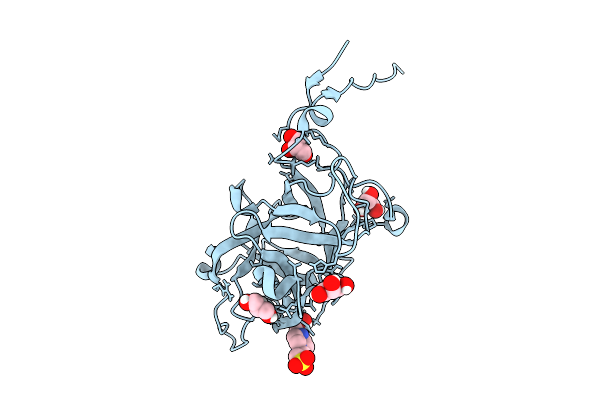 |
Structure Of The Saccharomyces Cerevisiae Pmt4-Mir Domain With Bound Ligands
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: PEPTIDE BINDING PROTEIN Ligands: EPE, GOL |
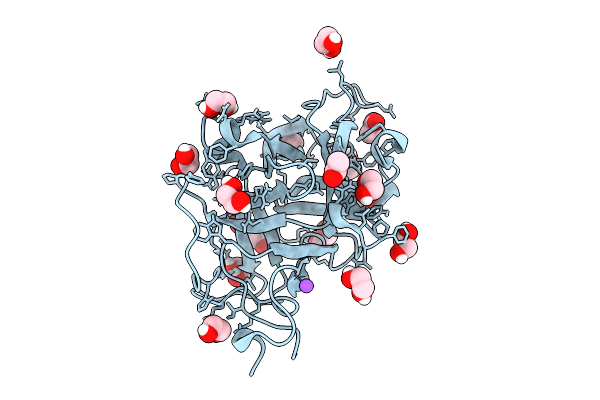 |
Structure Of The Chaetomium Thermophilum Pmt4-Mir Domain With Bound Ligands
Organism: Chaetomium
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, ACT, NA |
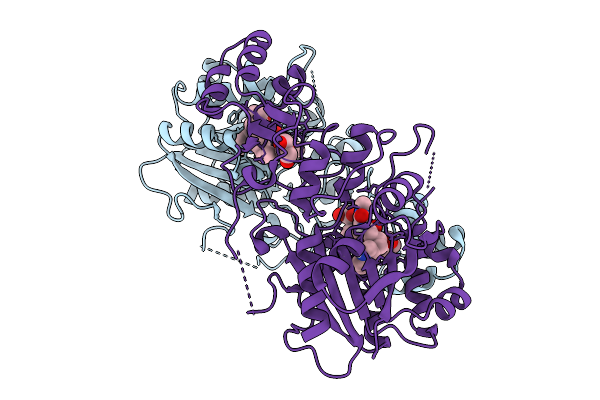 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: A1IUL, GOL |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: CHAPERONE Ligands: ATP, ADP |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: CHAPERONE Ligands: ATP, ADP |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: CHAPERONE Ligands: ATP, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RIBOSOME Ligands: GTP, GDP, MG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-09 Classification: RIBOSOME Ligands: MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: RIBOSOME Ligands: ZN, GTP, MG, GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: METAL BINDING PROTEIN Ligands: CU, EDO |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: MG, K, IHP, GTP, ZN |
 |
Organism: Schizosaccharomyces pombe
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: SPLICING Ligands: IHP, GTP, MG, ZN, K |
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa In A Detergent Bound Open Conformation
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE, PQE, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TRANSLATION Ligands: CO, IHP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: TRANSLATION Ligands: IHP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: METAL BINDING PROTEIN |
 |
Chaetomium Thermophilum Methionine Aminopeptidase 2 Autoproteolysis Product At The 80S Ribosome
Organism: Thermochaetoides thermophila dsm 1495, Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: METAL BINDING PROTEIN Ligands: ZN |

