Search Count: 258
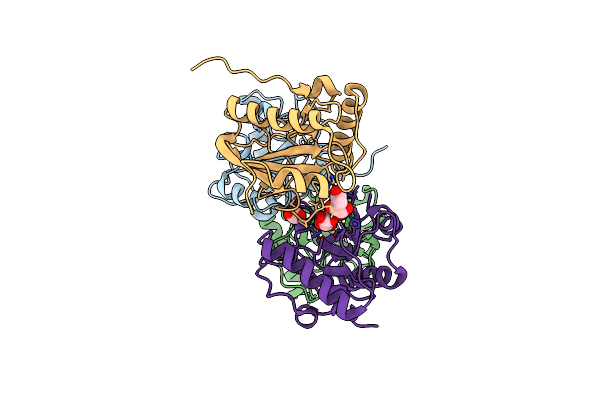 |
Crystal Structure Of The Complex Of Camel Peptidoglycan Recognition Protein, Pgrp-S With Malic Acid And Oxalic Acid At 2.3 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-05-14 Classification: IMMUNE SYSTEM Ligands: MLT, OXD |
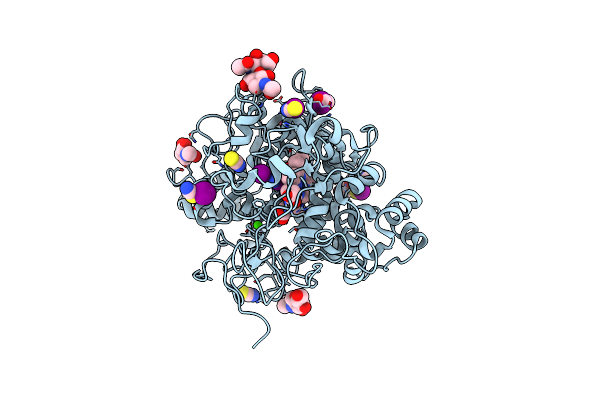 |
Structure Of The Intermediate Of Lactoperoxidase Formed With Thiocynate And Hydrogen Peroxidase At 1.99 A Resolution.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, SCN, PEO, GOL |
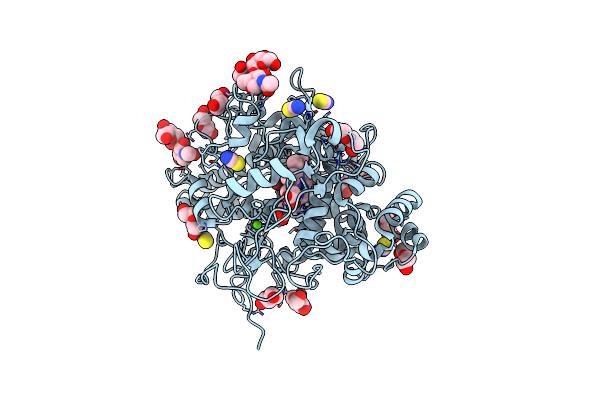 |
Crystal Structure Of The Complex Of Lactoperoxidase With Four Inorganic Substrates, Scn, I, Br And Cl
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: OXIDOREDUCTASE Ligands: HEM, CA, IOD, BR, CL, PEG, SCN, NAG |
 |
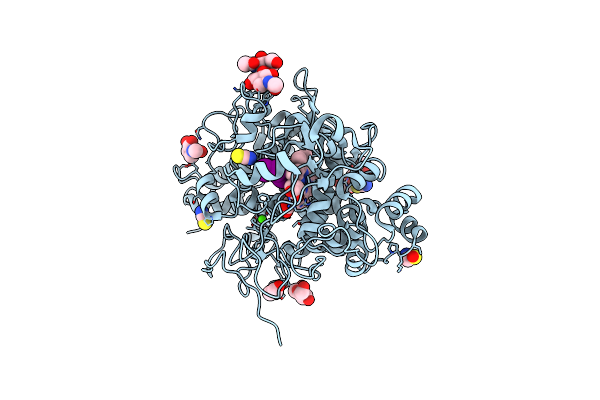
Structure Of The Ternary Complex Of Lactoperoxidase With Substrate Nitric Oxide (No) And Product Nitrite Ion (No2) At 1.98 A Resolution
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: NAG, NO, NO2, NO3, SCN, EDO, IOD, HEM, CA, OSM, NA |
 |
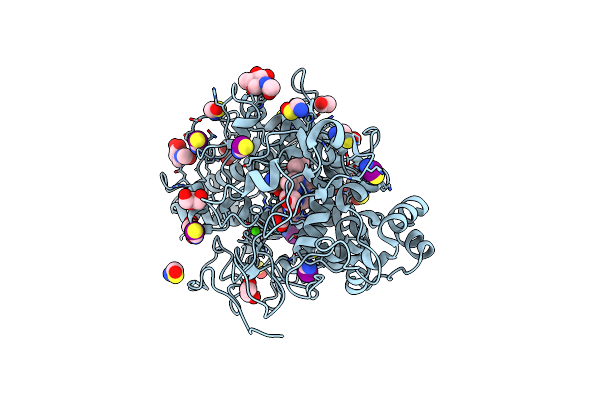
Structure Of Bovine Lactoperoxidase With Multiple Iodide Ions In The Distaline Heme Cavity.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2020-04-15 Classification: OXIDOREDUCTASE Ligands: CA, HEM, NAG, IOD, SCN, GOL, OSM, CL |
 |
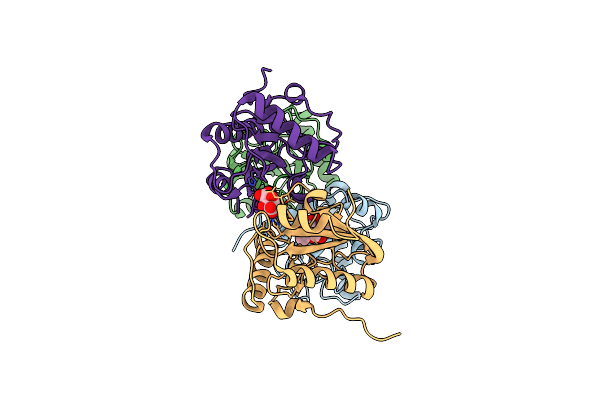
Crystal Structure Of The Complex Of Goat Lactoperoxidase With Hypothiocyanite And Hydrogen Peroxide At 1.79 A Resolution.
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-01-22 Classification: OXIDOREDUCTASE Ligands: NAG, HEM, CA, IOD, EDO, PEO, OSM, SCN, GOL |
 |
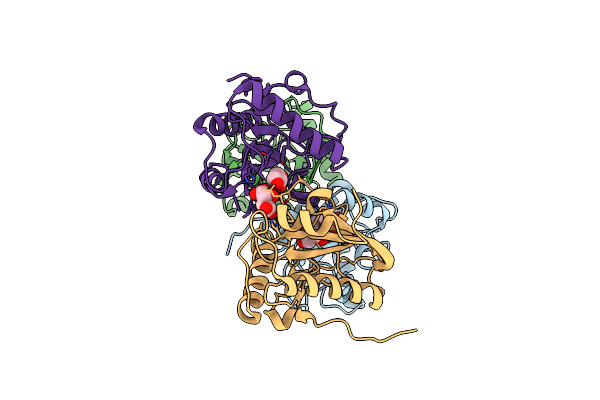
Crystal Structure Of The Complex Of Camel Peptidoglycan Recognition Protein (Cpgrp-S) And N-Acetylglucosamine At 2.6 A
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-10-14 Classification: IMMUNE SYSTEM Ligands: NAG, TLA |
 |
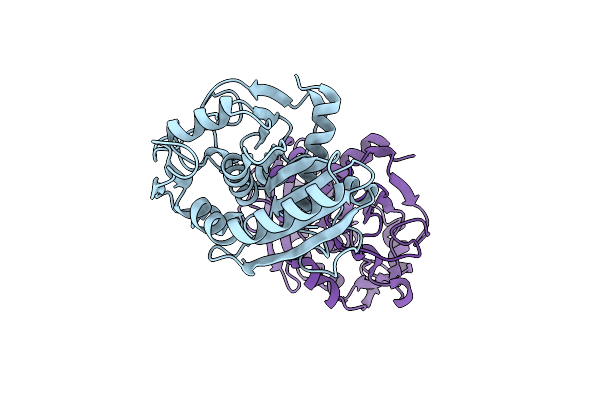
Crystal Structure Of The Complex Of Peptidoglycan Recognition Protein Pgrp-S With N-Acetyl Muramic Acid At 2.6 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-10-14 Classification: IMMUNE SYSTEM Ligands: TLA, AMU, GOL |
 |
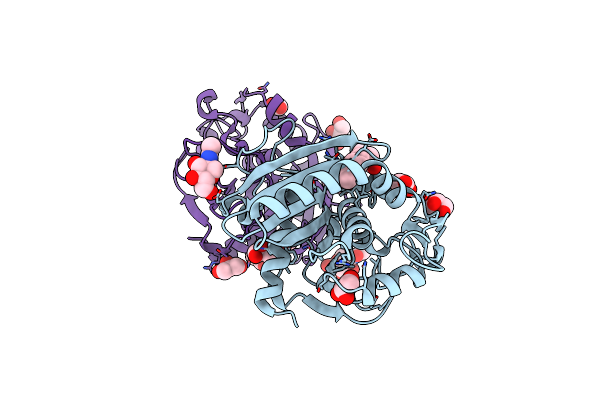
Structure Of Untreated Lipase From Thermomyces Lanuginosa At 2.3 A Resolution
Organism: Thermomyces lanuginosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-05-06 Classification: HYDROLASE |
 |
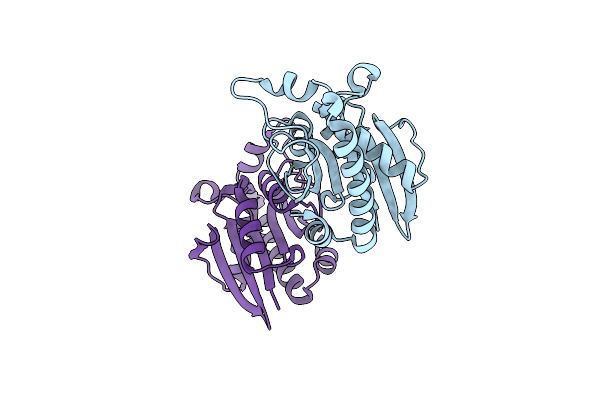
Structure Of Three Phase Partition - Treated Lipase From Thermomyces Lanuginosa In Complex With Lauric Acid At 2.1 A Resolution
Organism: Thermomyces lanuginosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: NAG, GOL, ACT, DAO, EDO, XXH |
 |
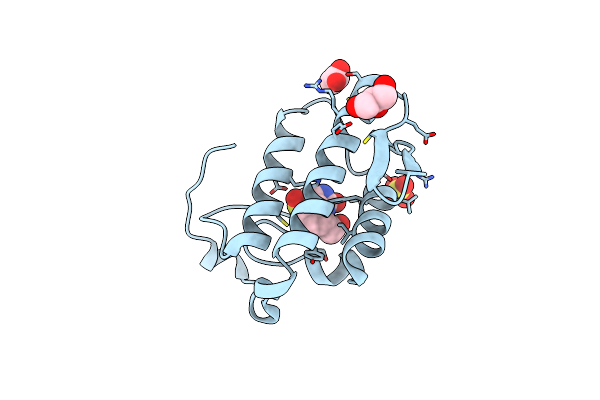
Crystal Structure Of Peptidyl-Trna Hydrolase From A Gram-Positive Bacterium, Streptococcus Pyogenes At 2.19 Angstrom Resolution Shows The Closed Structure Of The Substrate Binding Cleft
Organism: Streptococcus pyogenes nz131
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-08-06 Classification: HYDROLASE |
 |
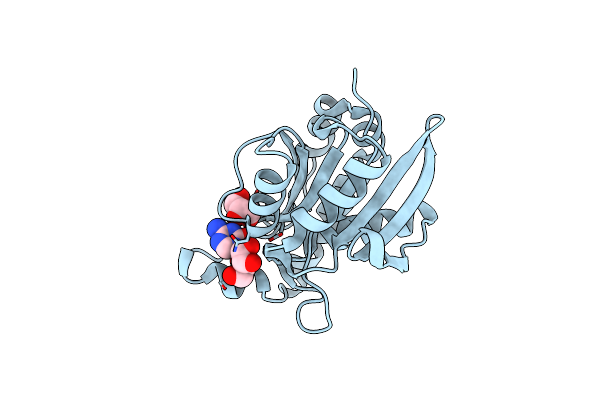
Crystal Structure Of Complex Formed Between Phospholipase A2 And Biotin-Sulfoxide At 1.09 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2014-07-30 Classification: HYDROLASE Ligands: GOL, BSO, SO4, ACT |
 |
Crystal Structure Of Peptidyl-Trna Hydrolase From Pseudomonas Aeruginosa With 5-Azacytidine At 1.89 Angstrom Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: 5AE, GOL |
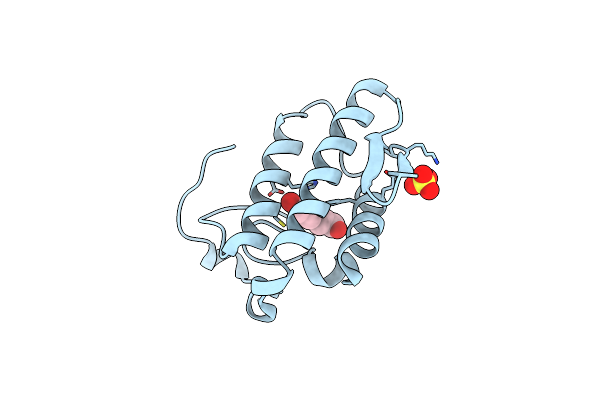 |
Crystal Structure Of The Complex Of Phospholipase A2 With P-Coumaric Acid At 1.2 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, HC4 |
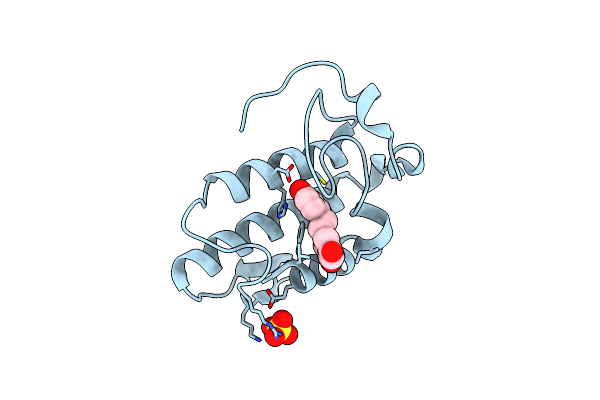 |
Crystal Structure Of The Complex Of Phospholipase A2 With Resveratrol At 1.20 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, STL |
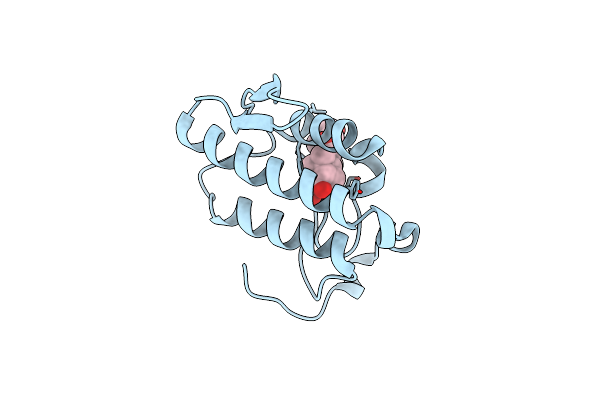 |
Crystal Structure Of The Complex Of Phospholipase A2 With Corticosterone At 1.48 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: C0R |
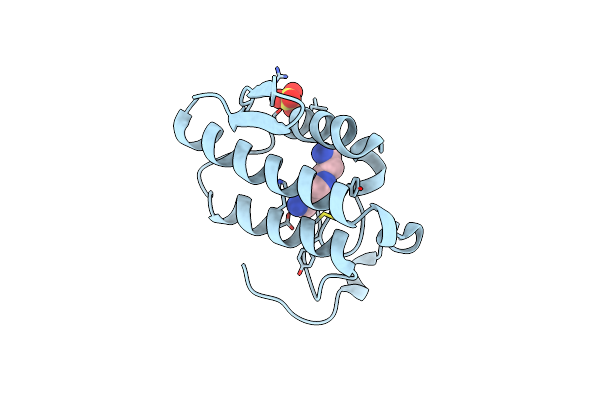 |
Crystal Structure Of The Complex Of Phospholipase A2 With Spermidine At 1.65 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SPD, SO4 |
 |
Crystal Structure Of The Complex Of Phospholipase A2 With Gramine Derivative At 1.80 A Resolution
Organism: Daboia russellii pulchella
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: PZZ |
 |
Crystal Structure Of Goat Lactoperoxidase In Complex With Octopamine At 2.1 Angstrom Resolution
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-06-18 Classification: OXIDOREDUCTASE Ligands: HEM, NAG, CA, IOD, SCN, OTR, PEG, EDO |
 |
Crystal Structure Of Type 1 Ribosome Inactivating Protein From Momordica Balsamina In Complex With Guanosine Mono Phosphate At 1.75 Angstrom Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: NAG, 5GP, GOL |

