Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
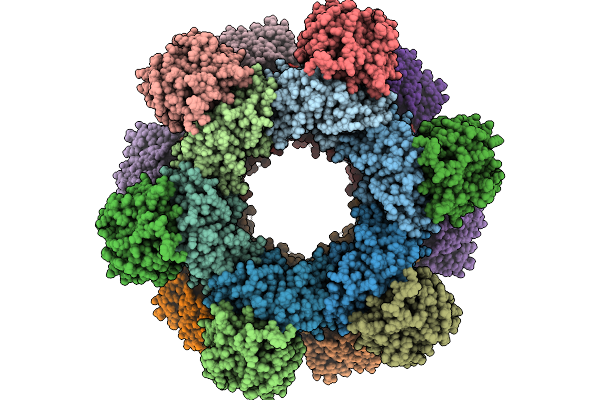 |
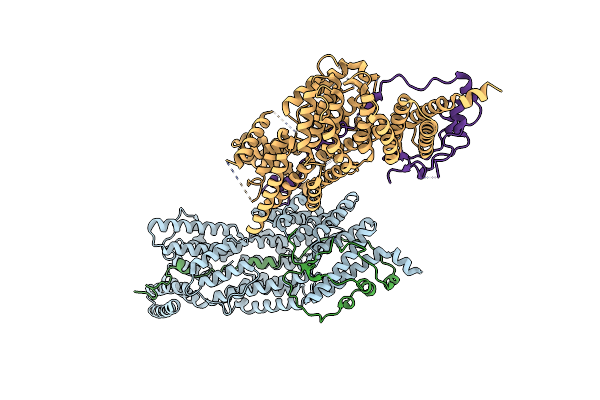
Cryo-Em Structure Of A Designed Pyridoxal Phosphate (Plp) Synthase Fused To A Designed Circumsporozoite Protein Antigen From Plasmodium Falciparum (Csp-P1-Csp And Csp-P2-Csp)
Organism: Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: BIOSYNTHETIC PROTEIN |
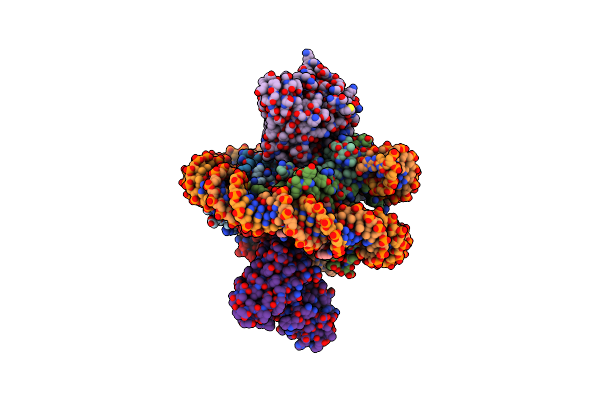 |
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN/DNA |
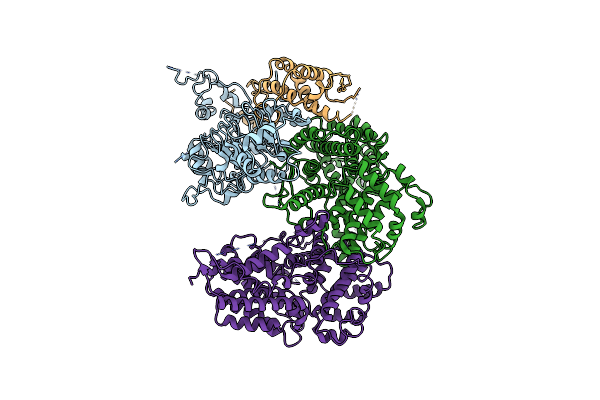 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN |
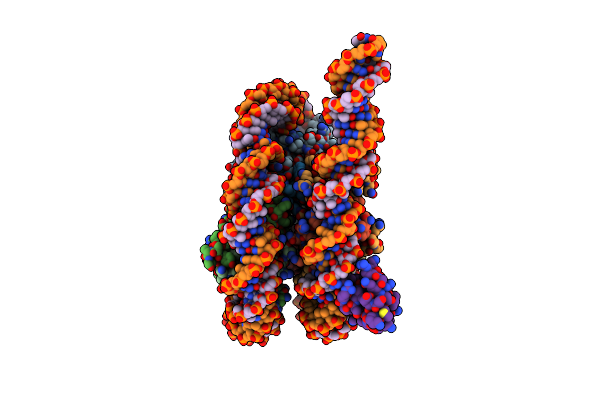 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-08-07 Classification: CELL CYCLE Ligands: BR |
 |
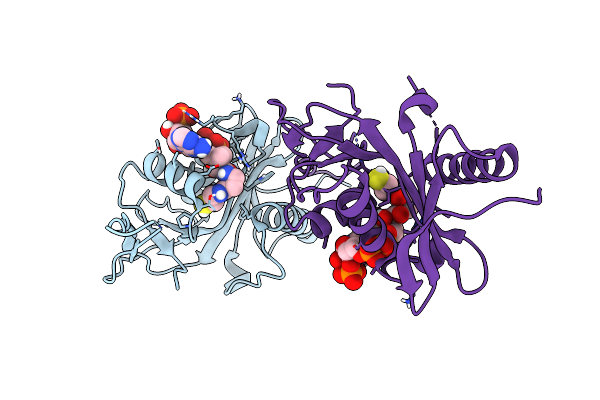
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2017-12-13 Classification: CELL CYCLE Ligands: ZN |
 |
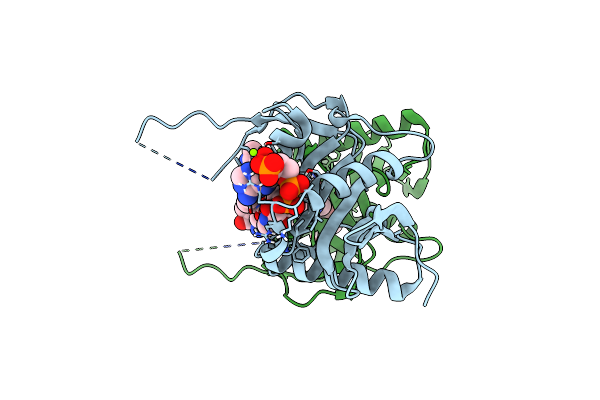
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2017-03-15 Classification: CELL CYCLE Ligands: COA |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-03-15 Classification: CELL CYCLE Ligands: MG, 8HB |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-03-15 Classification: CELL CYCLE Ligands: 8HB |
 |
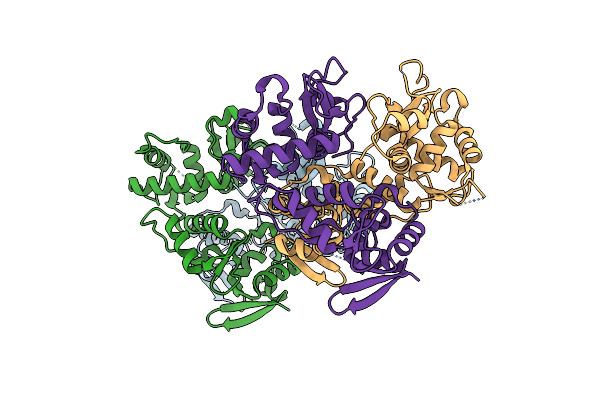
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-02-15 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-15 Classification: CELL CYCLE |
 |
Organism: Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056), Eremothecium gossypii atcc 10895
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-01-11 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2016-12-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe 972h-, Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-12-07 Classification: CELL CYCLE |
 |
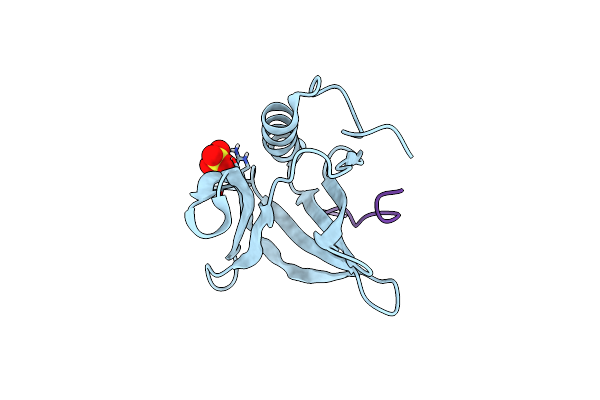
Organism: Ashbya gossypii, Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-08-05 Classification: CELL CYCLE |
 |
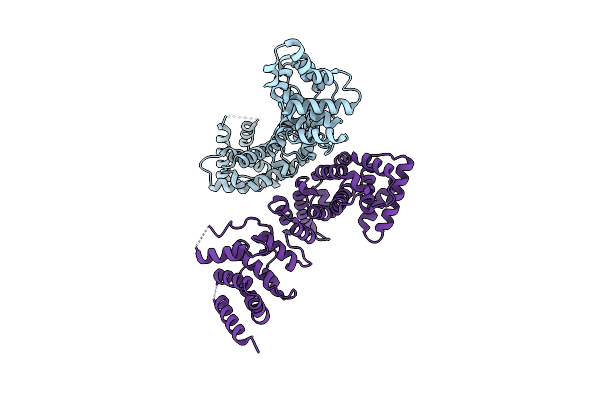
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
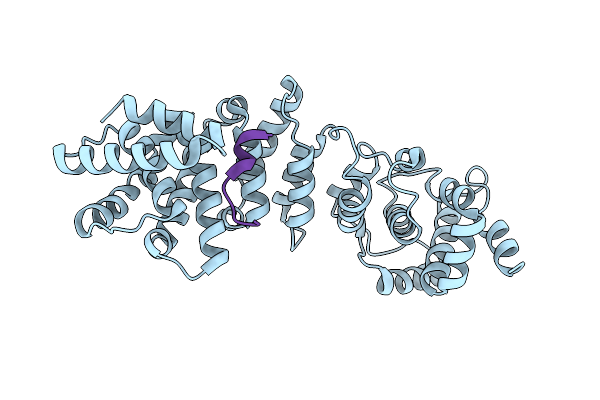
Organism: Eremothecium gossypii
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2013-02-20 Classification: CELL CYCLE |
 |
Organism: Eremothecium gossypii
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-02-20 Classification: CELL CYCLE |