Search Count: 650
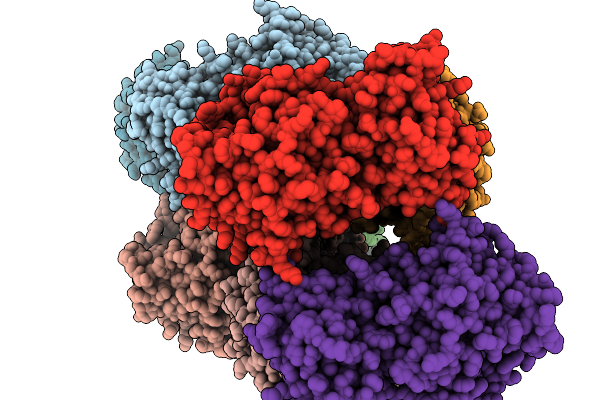 |
Crystal Structure Of Pseudopedobacter Saltans Gh43 Beta-Xylosidase In Complex With Xylose.
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2026-01-07 Classification: HYDROLASE Ligands: CA, XLS |
 |
Crystal Structure Of The Chymotrypsin-Cleaved Iron-Free C-Lobe Of Bovine Lactoferrin At 2.82 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-12-31 Classification: METAL BINDING PROTEIN Ligands: EDO, GOL, ACT, NAG, SO4 |
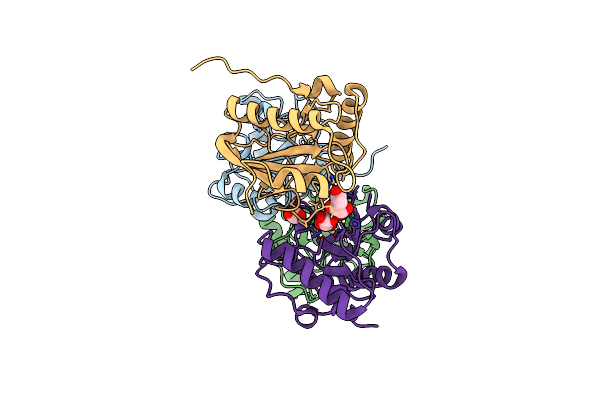 |
Crystal Structure Of The Complex Of Camel Peptidoglycan Recognition Protein, Pgrp-S With Malic Acid And Oxalic Acid At 2.3 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-05-14 Classification: IMMUNE SYSTEM Ligands: MLT, OXD |
 |
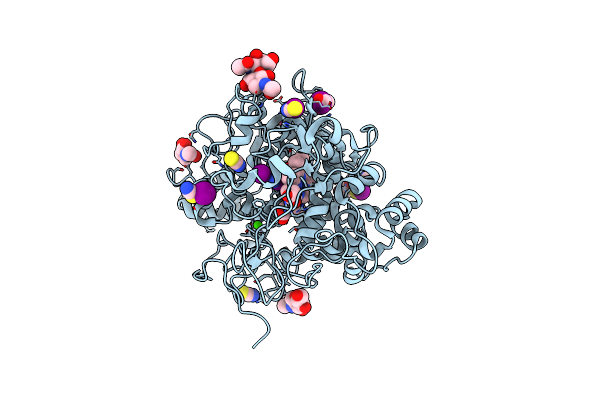
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The 17-Mer Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.10 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: CIT |
 |
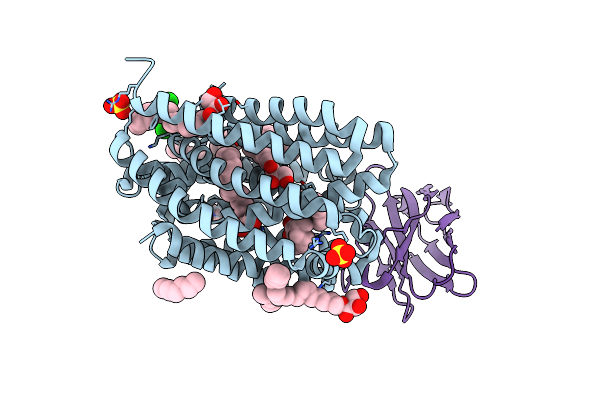
Structure Of The Intermediate Of Lactoperoxidase Formed With Thiocynate And Hydrogen Peroxidase At 1.99 A Resolution.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, SCN, PEO, GOL |
 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
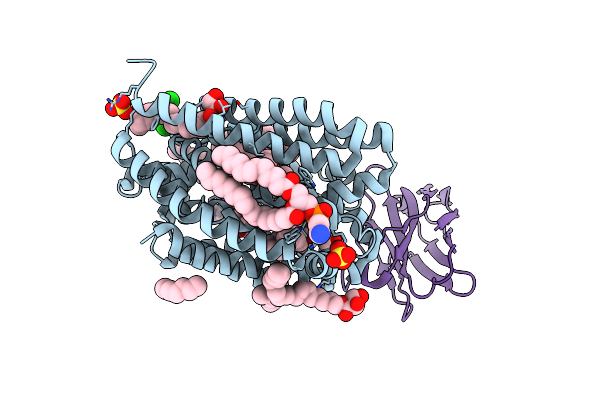 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
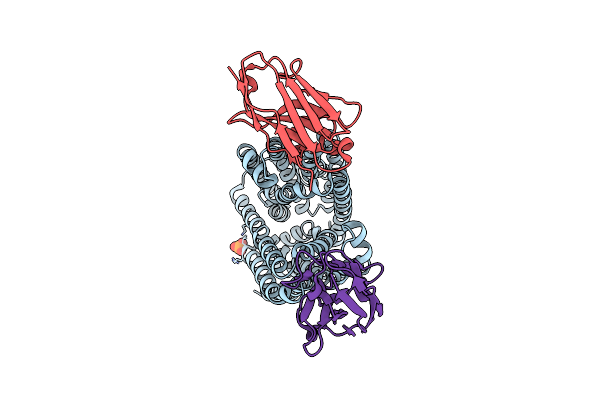 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: PO4 |
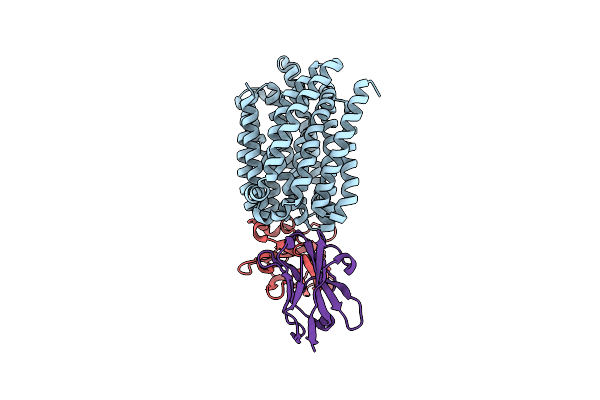 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 1
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
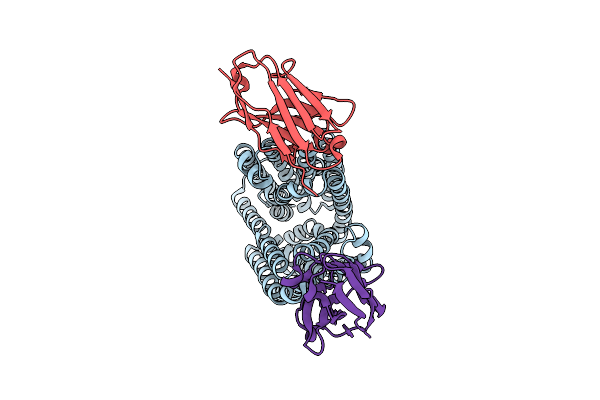 |
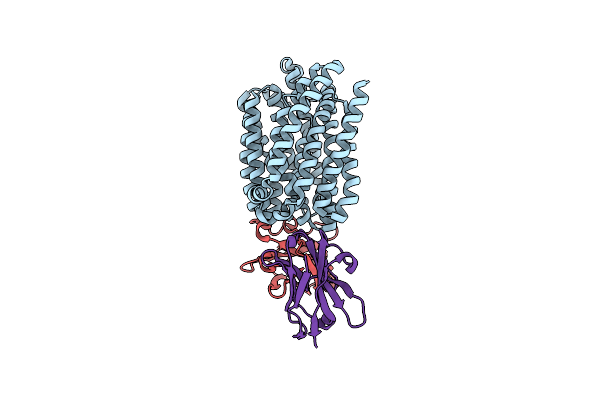
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Outward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Lipid Iii Flippase Wzxe With Nb10 And Nb7 Nanobodies In Inward-Facing Conformation - Crystal 2
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Nitric Oxide At 1.72 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: NAG, IOD, SCN, PGE, NO2, NO, HEM, CA |
 |
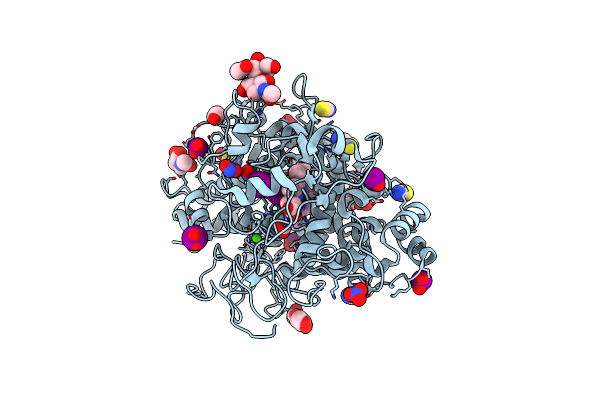
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.20 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: PAE, GOL |
 |
Crystal Structure Of The Ternary Complex Of Lactoperoxidase With Nitric Oxide And Nitrite Ion At 1.95 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE Ligands: NAG, NO, NO2, NO3, SCN, IOD, HEM, EDO, CA, OSM |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.39 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.37 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The 17-Mer Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.60 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: CIT, GOL |
 |
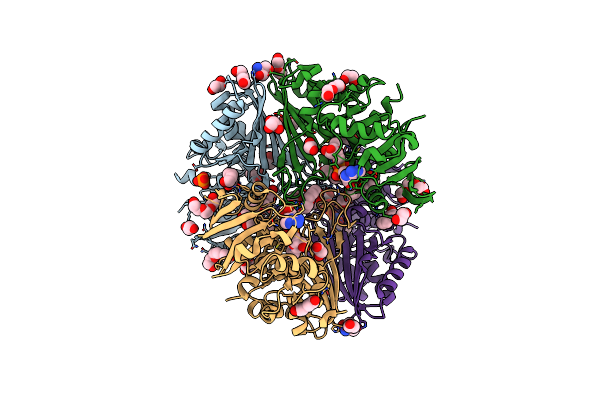
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.25 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
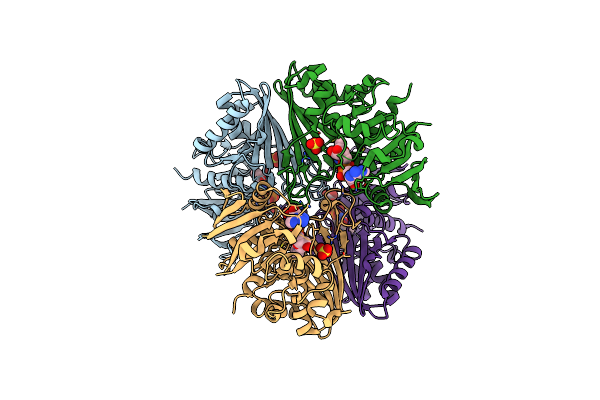
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide In The Presence Of Poly(Ethylene Glycol) At 2.20 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, PO4, PGE, TRS, 1PE, GOL |
 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide At 2.74 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |

