Search Count: 94
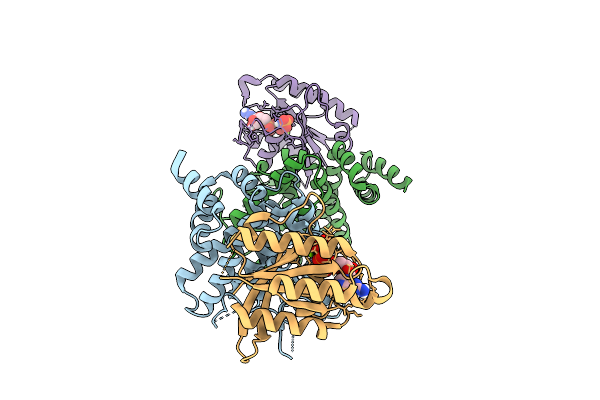 |
Crystal Structure Of The Human Ralgapa2 N-Terminal Domain With Human Kappab-Ras1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2025-07-16 Classification: SIGNALING PROTEIN Ligands: GNP, MG |
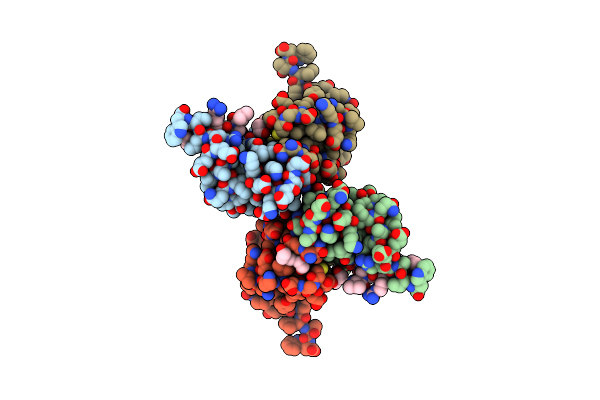 |
Crystal Structure Of Hp1Alpha Chromoshadow Domain In Complex With Kap1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-06-11 Classification: TRANSCRIPTION/PEPTIDE Ligands: PGE |
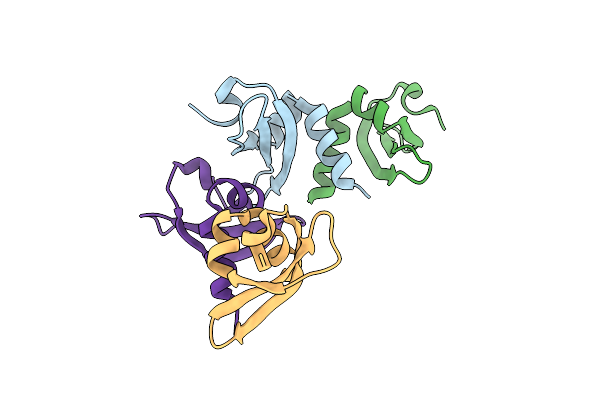 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-06-11 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-08-17 Classification: TRANSCRIPTION |
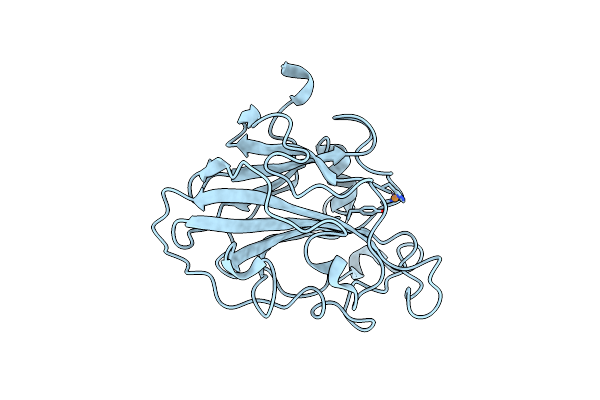 |
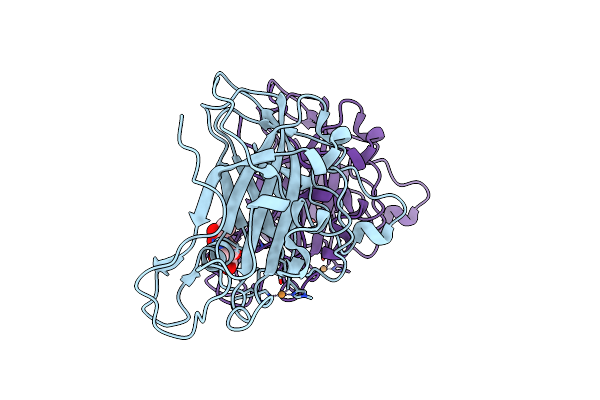
High Resolution X-Ray Structure Of E. Coli Expressed Lentinus Similis Lpmo.
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-03-16 Classification: CARBOHYDRATE Ligands: CU, CL |
 |
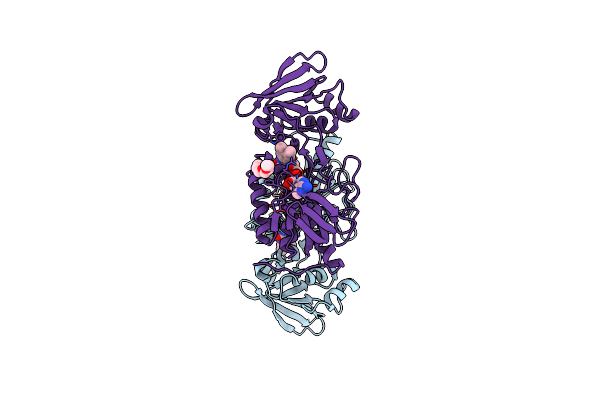
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-03-16 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
 |
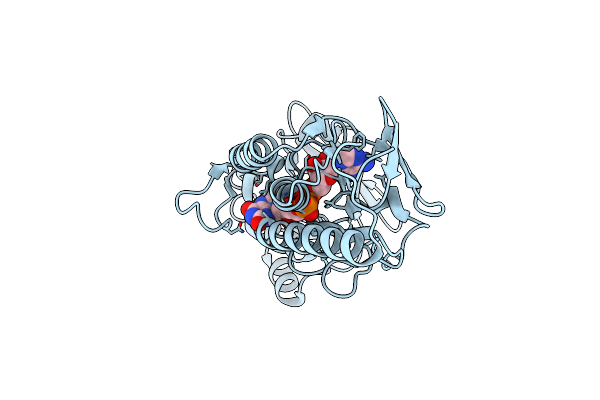
Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain Cs Mutant
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2022-01-19 Classification: OXIDOREDUCTASE Ligands: FAD, P4G |
 |
Crystal Structure Of Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FAD |
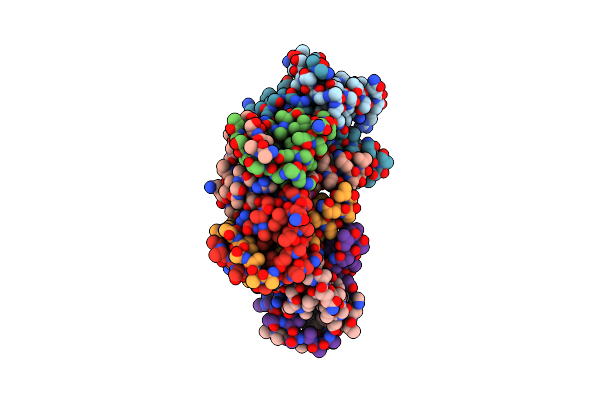 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2021-10-06 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis In Complex With Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: 3PO, MG, EU9 |
 |
Crystal Structure Of Apo Form Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: MG, PO4 |
 |
Acel Nrdhf Class 3 Split Intein Gsh Linked Splice Inactive Variant - C124A, N146A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-16 Classification: SPLICING Ligands: IOD |
 |
Crystal Structure Of The Lrr-Roc-Cor Domain Of The Chlorobium Tepidum Roco Protein
Organism: Chlorobaculum tepidum
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2018-12-26 Classification: HYDROLASE |
 |
Crystal Structure Of Wild Type Phosphoserine Aminotransferase (Psat) From E. Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2018-10-31 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Crystal Structure Of Delta 4 Mutant Of Ehpsat (Phosphoserine Aminotransferase Of Entamoeba Histolytica)
Organism: Entamoeba histolytica hm-3:imss
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-10-31 Classification: TRANSFERASE Ligands: CL, PLP |
 |
Crystal Structure Of 45 Amino Acid Deleted From N-Terminal Of Phosphoserine Aminotransferase (Psat) Of Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: SCN |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: ACT, GOL, TRS |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: SAM, GOL |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: SAH |

