Search Count: 191
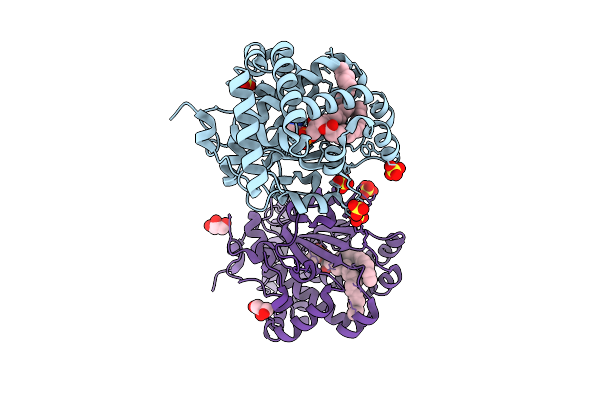 |
Crystal Structure Of Saccharomyces Cerevisiae Cross-Linked Sfh1 (K197C, F233C) Mutant In Complex With Phosphatidylethanolamine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: PTY, SO4, GOL |
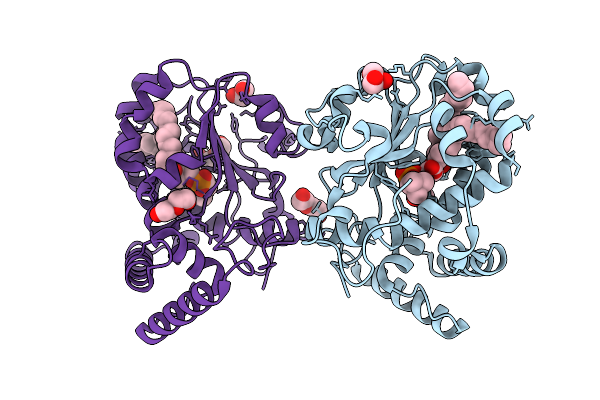 |
Crystal Structure Of Saccharomyces Cerevisiae Sec14P In Complex With Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, GOL, EDO, PEG |
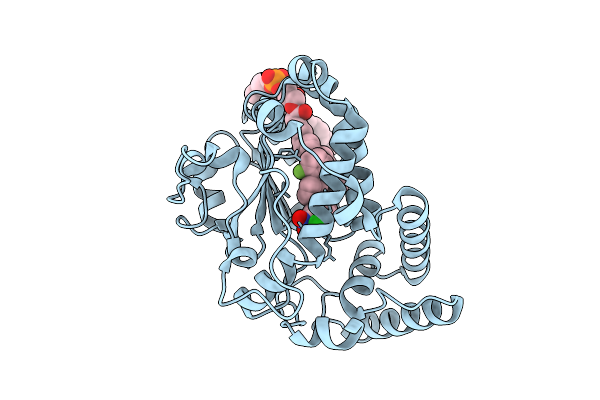 |
Crystal Structure Of A Ternary Complex Of Saccharomyces Cerevisiae Sec14 With A Small Molecule Inhibitor And Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, IUO |
 |
Cryo-Em Structure Of Amyloid Fibril Extracted From Heart Of A Variant Attr T60A Amyloidosis Patient 1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Amyloid Fibril Extracted From Heart Of A Variant Attr T60A Amyloidosis Patient 2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Amyloid Fibril Extracted From Thyroid Of A Variant Attr T60A Amyloidosis Patient 3
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Amyloid Fibril Extracted From Kidney Of A Variant Attr T60A Amyloidosis Patient 3
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Amyloid Fibril Extracted From Liver Of A Variant Attr T60A Amyloidosis Patient 3
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: PROTEIN FIBRIL |
 |
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cec215
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 1PE, SO4, A1ATV |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERSE INHIBITOR Ligands: 1PE, SO4, A1ATW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: 1PE, SO4 |
 |
M. Tuberculosis Pks13 Acyltransferase (At) Domain In Complex With Sufex Inhibitor Cmx410 - Reaction Product
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: SO4, 1PE, A1AVL |
 |
Ccw Flagellar Switch Complex - Flif, Flig, Flim, And Flin Forming 34-Mer C-Ring From Salmonella
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MOTOR PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN FIBRIL Ligands: NAG |
 |
C-Ring - Single Subunit Of The 34-Mer Ccw Flagellar Switch Complex - Flif, Flig, Flim, And Flin From Salmonella
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 Mutant From Uncultured Bacterium In Complex With Ligand
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: A1D5H, GOL |
 |
Crystal Structure Of A Novel Pu Plastic Degradation Urethanase Umg-Sp2 From Uncultured Bacterium
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, PMS |

