Search Count: 62
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-04-23 Classification: TOXIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-04-23 Classification: TOXIN |
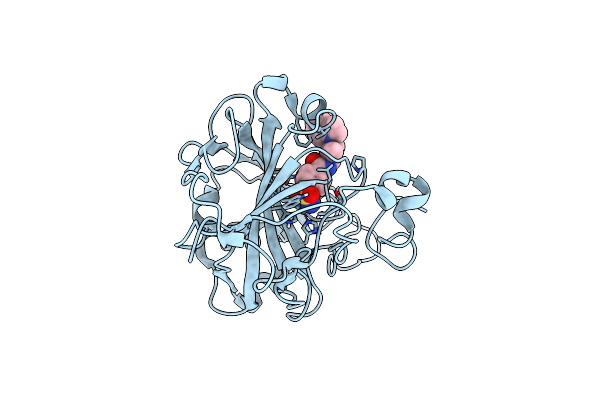 |
Human Carbonic Anhydrase Ii (Hcaii) In Complex With (R)-N-(3-Indol-1-Yl-2-Methyl-Propyl)-4-Sulfamoyl-Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-12-06 Classification: METAL BINDING PROTEIN Ligands: SBR, ZN |
 |
The Solution Structure Of The Triple Mutant Methyl-Cpg-Binding Domain From Mecp2
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-22 Classification: GENE REGULATION |
 |
The Solution Structure Of The Triple Mutant Methyl-Cpg-Binding Domain From Mecp2 That Binds To Asymmetrically Modified Dna
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-22 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2021-11-03 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: V84 |
 |
Organism: Sus scrofa
Method: SOLUTION NMR Release Date: 2020-08-12 Classification: PEPTIDE BINDING PROTEIN |
 |
Active-Site Conformational Dynamics Of Carbonic Anhydrase Ii Under Native Conditions: An Nmr Perspective
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-08-28 Classification: HYDROLASE Ligands: ZN |
 |
Assessment Of A Large Enzyme-Drug Complex By Proton-Detected Solid-State Nmr Without Deuteration
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2019-02-06 Classification: HYDROLASE Ligands: ZN, AZM |
 |
Organism: Chlamydomonas reinhardtii
Method: SOLUTION NMR Release Date: 2018-10-03 Classification: PLANT PROTEIN |
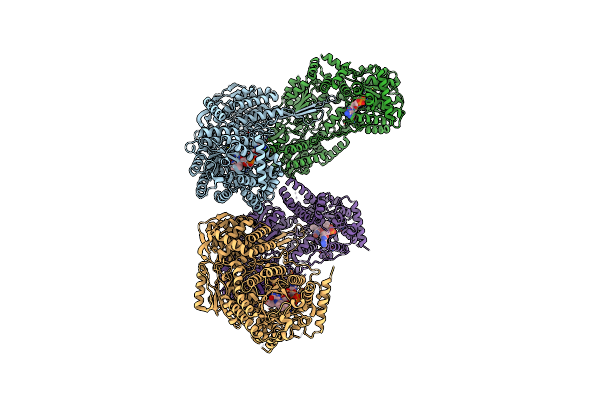 |
Crystal Structure Of Proline Utilization A (Puta) From Bdellovibrio Bacteriovorus Inactivated By N-Propargylglycine
Organism: Bdellovibrio bacteriovorus (strain atcc 15356 / dsm 50701 / ncib 9529 / hd100)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-08-02 Classification: OXIDOREDUCTASE Ligands: P5F |
 |
Crystal Structure Of Oxidized Aspergillus Fumigatus Udp-Galactopyranose Mutase Complexed With Nadph
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-05-31 Classification: ISOMERASE Ligands: FAD, SO4, NDP |
 |
Crystal Structure Of Oxidized Aspergillus Fumigatus Udp-Galactopyranose Mutase Complexed With Nadh
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-05-31 Classification: ISOMERASE Ligands: FAD, NAI, SO4 |
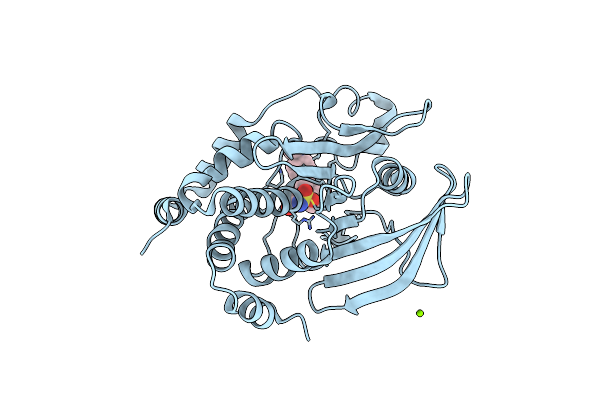 |
Structure Of Ptp1B Complexed With N-(3'-(1,1-Dioxido-4-Oxo-1,2,5-Thiadiazolidin-2-Yl)-4'-Methyl-[1,1'-Biphenyl]-4-Yl)Acetamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-04-12 Classification: HYDROLASE Ligands: MG, 73U, CL |
 |
Crystal Structure Of Nsp2 Protease From Chikungunya Virus In P212121 Space Group At 2.59 A (4Molecules/Asu).
Organism: Chikungunya virus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-06-15 Classification: HYDROLASE Ligands: GOL |
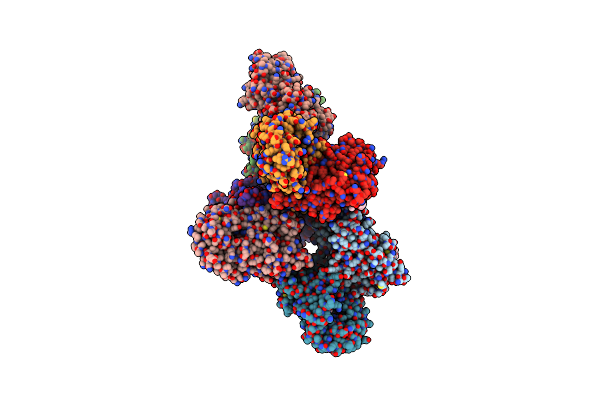 |
Structure Of Legionella Pneumophila Histidine Acid Phosphatase Complexed With L(+)-Tartrate
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-27 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TLA, 1PE |
 |
Nmr Solution Structure Of S114A Mutant Of A Uv Inducible Protein From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: SOLUTION NMR Release Date: 2015-12-16 Classification: HYDROLASE INHIBITOR |
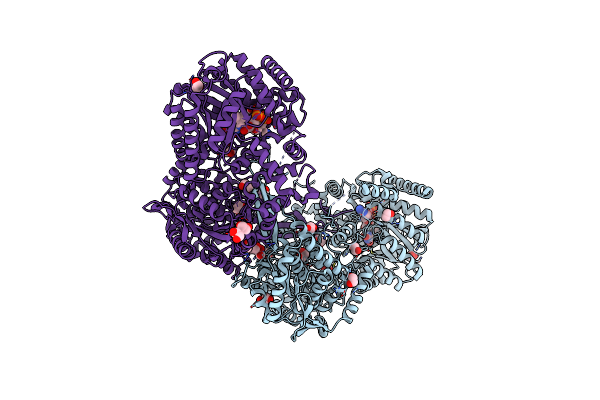 |
Crystal Structure Of The Resting State Of Proline Utilization A (Puta) From Geobacter Sulfurreducens Pca
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, EDO |
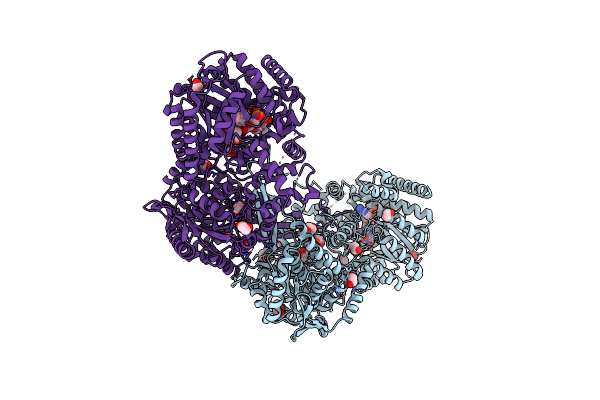 |
Crystal Structure Of Proline Utilization A (Puta) From Geobacter Sulfurreducens Pca In Complex With L-Tetrahydro-2-Furoic Acid
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, TFB, EDO |
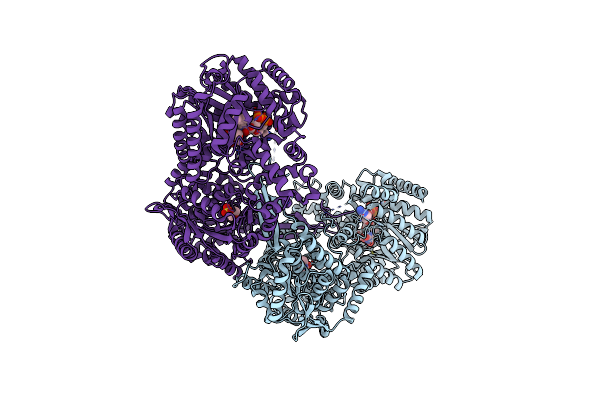 |
Crystal Structure Of Proline Utilization A (Puta) From Geobacter Sulfurreducens Pca In Complex With L-Lactate
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, 2OP, MES |

