Search Count: 30
 |
Crystal Structure Of Active Kras-G12C (Gmppnp-Bound) In Complex With Bbo-8520
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GNP, MG, Y8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, Y8N |
 |
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE |
 |
Structure Of E. Coli Dgtpase Bound To T7 Bacteriophage Protein Gp1.2 And Dgtp
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE Ligands: MG, DGT |
 |
Structure Of E. Coli Dgtpase Bound To T7 Bacteriophage Protein Gp1.2 And Gtp
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE Ligands: GTP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Crystal Structure Of Se-Met Labelled Mce Domain Of Mce4A From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2021-08-25 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Mce Domain Of Mce4A From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-08-25 Classification: TRANSPORT PROTEIN |
 |
Crystal Structures Of Beta-1,4-N-Acetylglucosaminyltransferase 2 (Pomgnt2): Structural Basis For Inherited Muscular Dystrophies
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: UDP, NAG, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: NAG, PO4 |
 |
Organism: Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2020-02-26 Classification: PROTEIN BINDING |
 |
Organism: Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2020-02-26 Classification: PROTEIN BINDING |
 |
Organism: Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2020-02-26 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2019-08-28 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-08-28 Classification: CELL CYCLE |
 |
Organism: Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2019-07-24 Classification: PROTEIN BINDING |
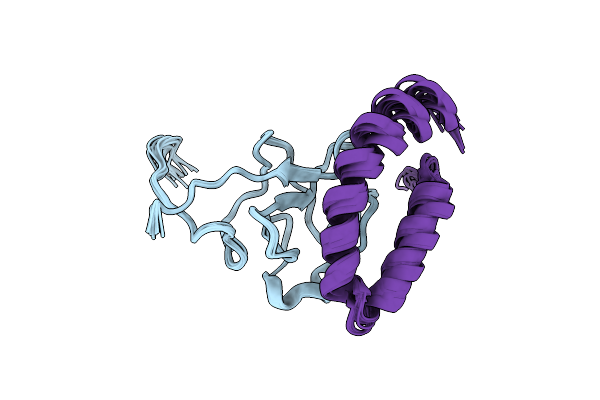 |
Solution Structure Of The Complex Of Mutant Vek50[Rh1/Aa] And Plasminogen Kringle 2
Organism: Homo sapiens, Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2019-07-24 Classification: PROTEIN BINDING |
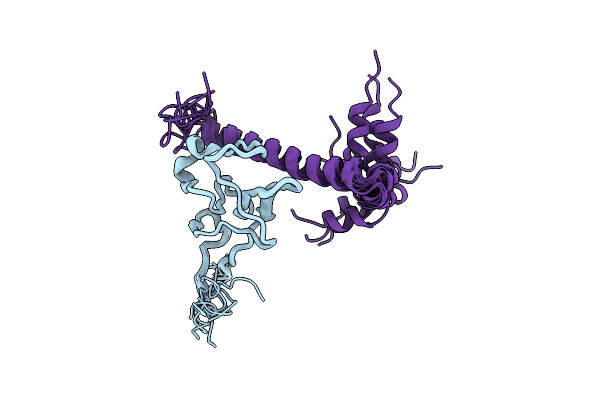 |
Solution Structure Of The Complex Of Mutant Vek50[Rh2/Aa] And Plasminogen Kringle 2
Organism: Homo sapiens, Streptococcus pyogenes
Method: SOLUTION NMR Release Date: 2019-07-24 Classification: PROTEIN BINDING |

