Search Count: 61
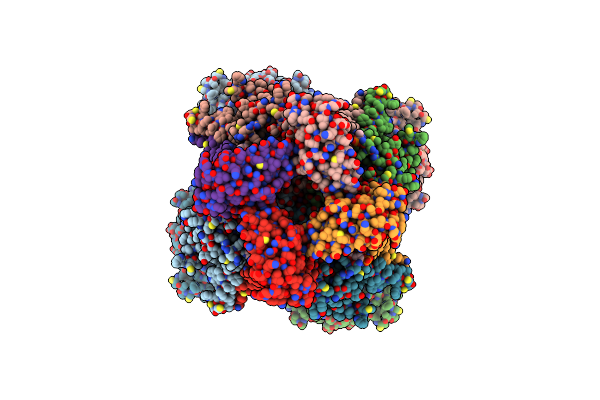 |
Ca2+ Bound Intermediate State Of Hslo1 + Beta2N-Beta4 Channel In Detergent.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA, K, POV |
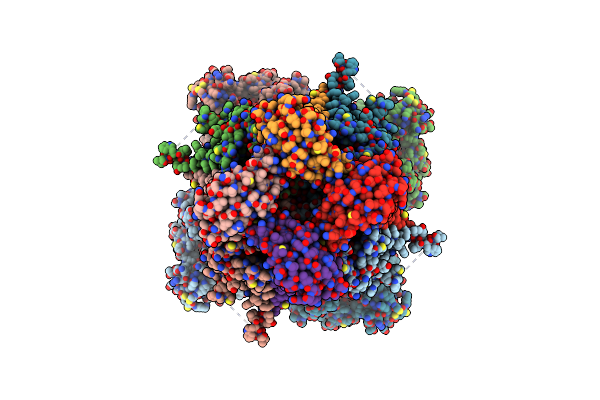 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CLR, K |
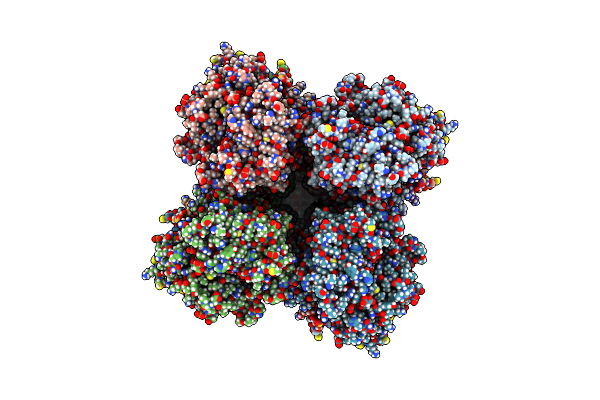 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: K, POV, CLR |
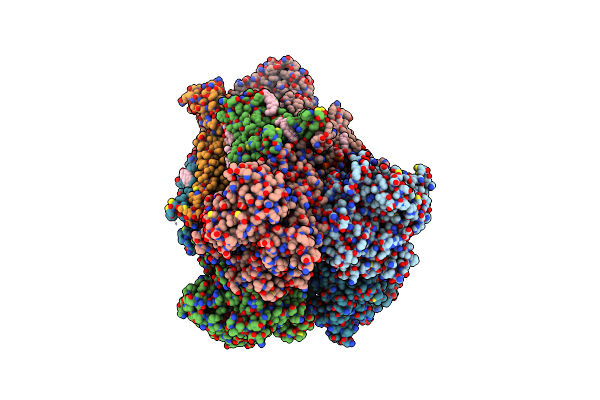 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: POV, MG, CA, CLR, K |
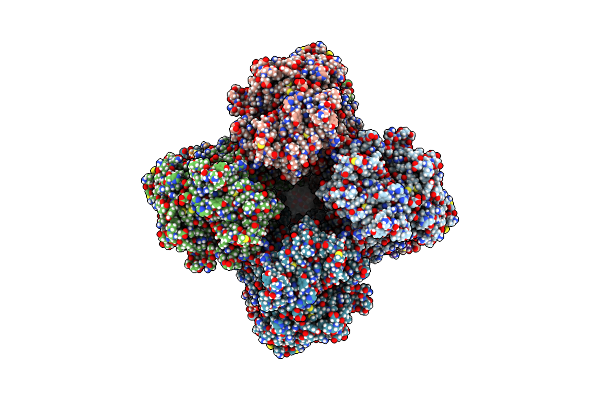 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: CA, K, POV |
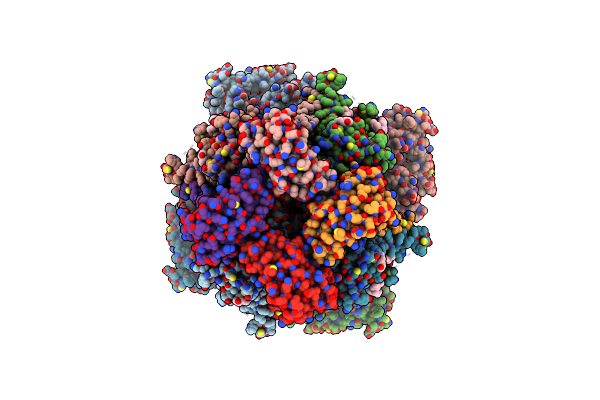 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: MG, CA, POV, CLR, K |
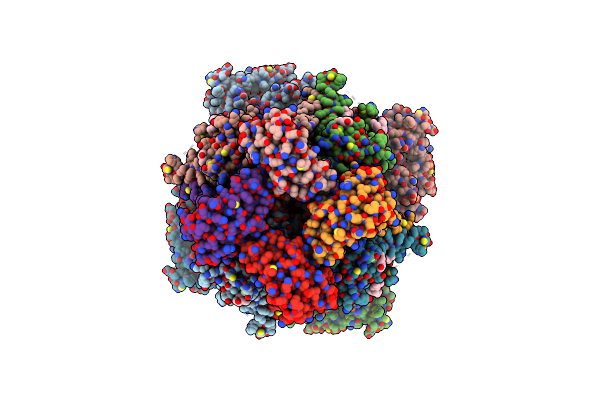 |
Ca2+ Bound Open-Inactivated Hslo1 + Beta2N-Beta4 Channel In Detergent-Conformation 2 Of Inactivating Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: MG, CA, POV, CLR, K |
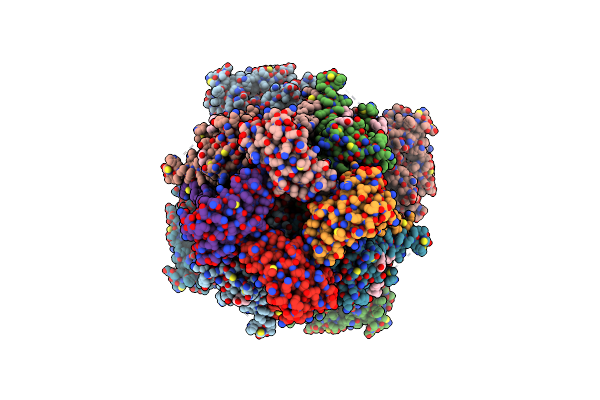 |
Ca2+ Bound Open-Inactivated Hslo1 + Beta2N-Beta4 Channel In Detergent-Conformation 3 Of Inactivating Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: MG, CA, POV, CLR, K |
 |
3D Cryo-Em Reveals The Structure Of A 3-Fmoc Zipper Motif Ensuring The Self-Assembly Of Tripeptide Nanofiber
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Structure Of Glutamate Receptor-Like Channel Glr3.4 Ligand-Binding Domain In Complex With Glutamate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: GLU, CL, SO4, GOL |
 |
Structure Of Glutamate Receptor-Like Channel Glr3.4 Ligand-Binding Domain In Complex With Serine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: SER, GOL, SO4 |
 |
Structure Of Glutamate Receptor-Like Channel Glr3.4 Ligand-Binding Domain In Complex With Methionine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: MET, NA, SO4, GOL, BME, CL |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: TRANSPORT PROTEIN Ligands: GLU, NAG, GSH |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: TRANSPORT PROTEIN Ligands: GLU |
 |
Structure Of The Glutamate-Like Receptor Glr3.2 Ligand-Binding Domain In Complex With Methionine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-09-23 Classification: MEMBRANE PROTEIN Ligands: MET, GOL, BME, CL |
 |
Structure Of The Glutamate-Like Receptor Glr3.2 Ligand-Binding Domain In Complex With Glycine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-09-23 Classification: MEMBRANE PROTEIN Ligands: GLY, NA, BME |
 |
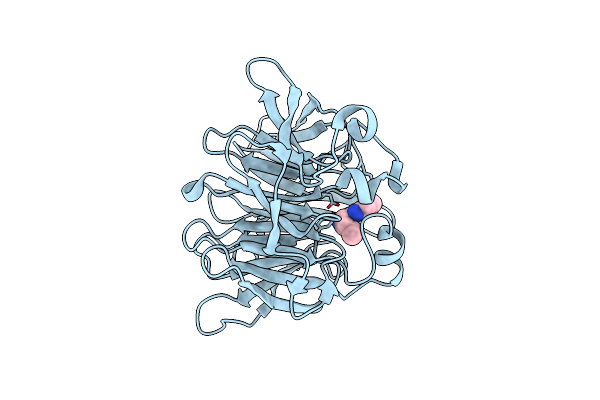
Strictosidine Synthase From Ophiorrhiza Pumila In Complex With (S)-1-Ethyl-2,3,4,9-Tetrahydro-1H-Beta-Carboline
Organism: Ophiorrhiza pumila
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2020-04-08 Classification: LYASE Ligands: KW8 |
 |
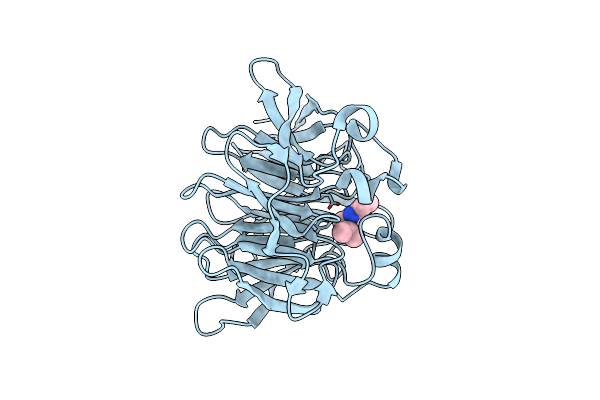
Strictosidine Synthase From Ophiorrhiza Pumila In Complex With (S)-1-N-Propyl-2,3,4,9-Tetrahydro-1H-Beta-Carboline
Organism: Ophiorrhiza pumila
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-04-08 Classification: LYASE Ligands: KWH |
 |
Strictosidine Synthase From Ophiorrhiza Pumila In Complex With (S)-1-Isobutyl-2,3,4,9-Tetrahydro-1H-Beta-Carboline
Organism: Ophiorrhiza pumila
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-04-08 Classification: LYASE Ligands: KWQ |

