Search Count: 12
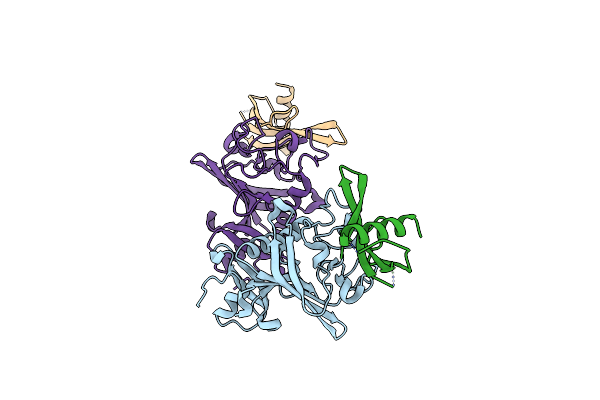 |
Crystal Structure Of The C-Terminal Domain Of Human Tnc In Complex With Adhiron 52
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: SIGNALING PROTEIN |
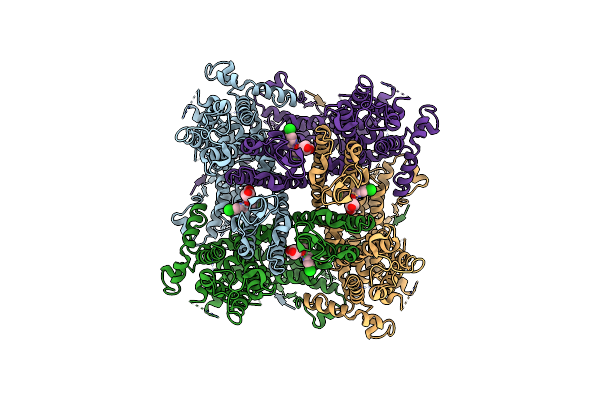 |
Organism: Escherichia coli (strain k12), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN Ligands: PJQ |
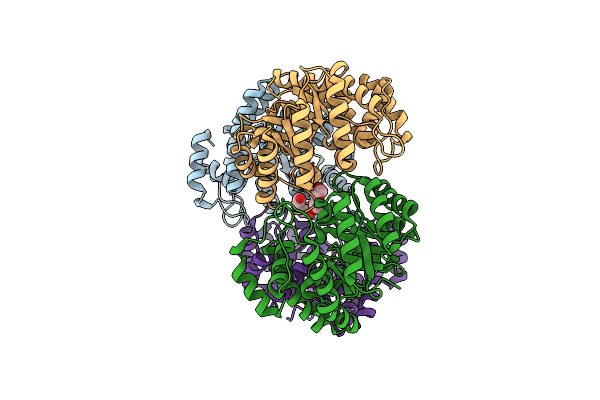 |
X-Ray Crystal Structure Of N-Acetylneuraminic Acid Lyase In Complex With Pyruvate, With The Phenylalanine At Position 190 Replaced With The Non-Canonical Amino Acid Dihydroxypropylcysteine.
Organism: Staphylococcus aureus (strain nctc 8325)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-22 Classification: LYASE Ligands: PEG |
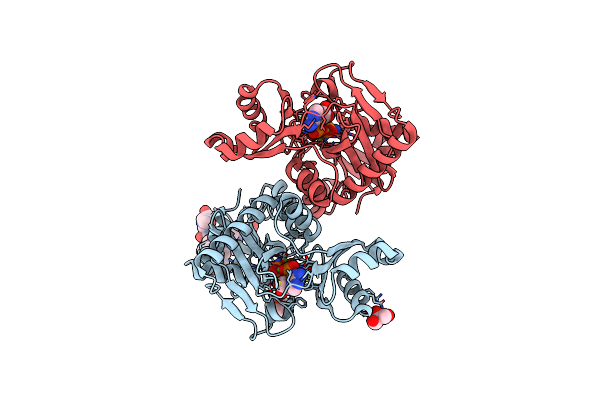 |
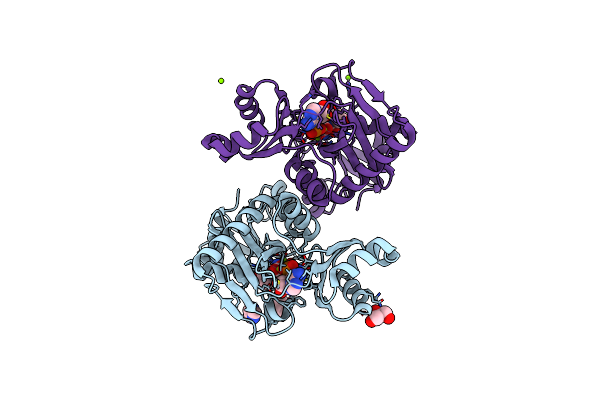
The X-Ray Crystal Structures Of D-Alanyl-D-Alanine Ligase In Complex Adp And D-Cycloserine Phosphate
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-01-21 Classification: LIGASE Ligands: ADP, DS0, MG, GOL |
 |
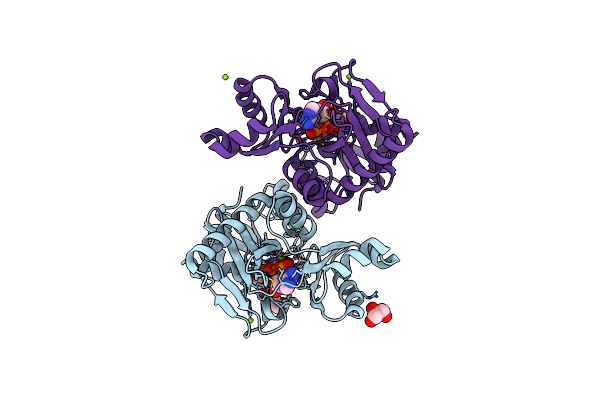
The X-Ray Crystal Structure Of D-Alanyl-D-Alanine Ligase In Complex With Atp And D-Ala-D-Ala
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-01-21 Classification: LIGASE Ligands: ADP, DAL, CO3, MG, GOL, IMD |
 |
The X-Ray Crystal Structure Of D-Alanyl-D-Alanine Ligase In Complex With Adp And D-Ala-D-Ala
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-01-21 Classification: LIGASE Ligands: ATP, DAL, MG, GOL |
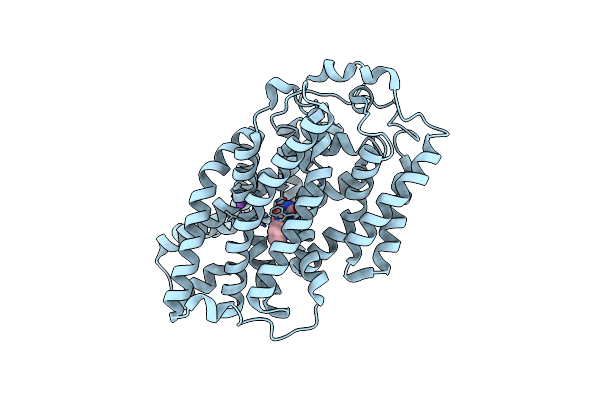 |
Structure Of Mhp1, A Nucleobase-Cation-Symport-1 Family Transporter, In A Closed Conformation With Indolylmethyl-Hydantoin
Organism: Microbacterium liquefaciens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-07-02 Classification: TRANSPORT PROTEIN Ligands: I5H, NA |
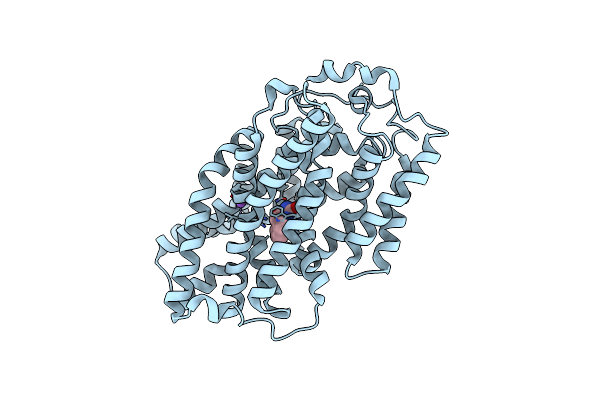 |
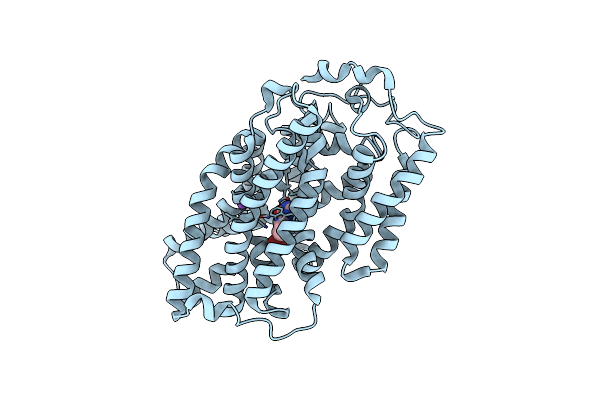
Structure Of Mhp1, A Nucleobase-Cation-Symport-1 Family Transporter, In A Closed Conformation With Benzyl-Hydantoin
Organism: Microbacterium liquefaciens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2014-07-02 Classification: TRANSPORT PROTEIN Ligands: 5FH, NA |
 |
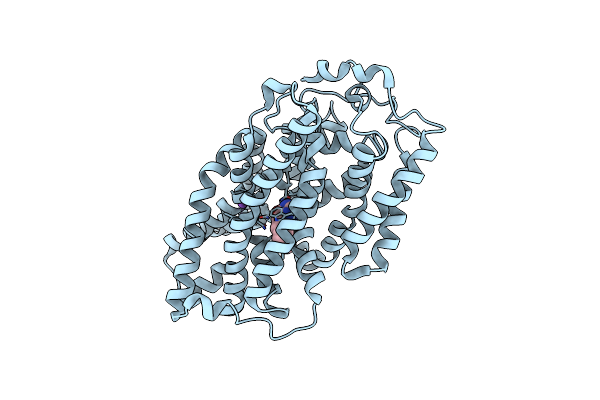
Structure Of Mhp1, A Nucleobase-Cation-Symport-1 Family Transporter, In A Closed Conformation With Bromovinylhydantoin Bound.
Organism: Microbacterium liquefaciens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2014-07-02 Classification: TRANSPORT PROTEIN Ligands: B5H, NA |
 |
Structure Of Mhp1, A Nucleobase-Cation-Symport-1 Family Transporter With The Inhibitor 5-(2-Naphthylmethyl)-L-Hydantoin.
Organism: Microbacterium liquefaciens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2014-07-02 Classification: TRANSPORT PROTEIN Ligands: 5NL, 5ND, NA |
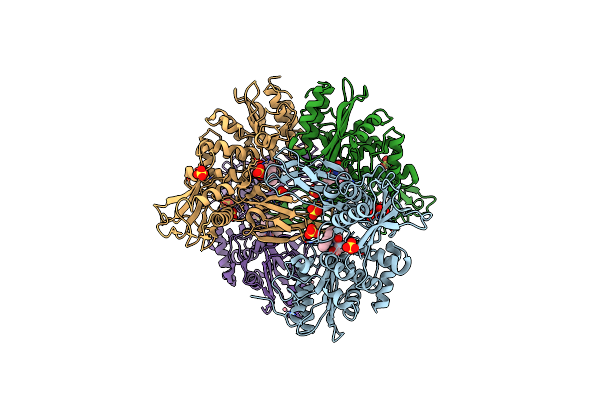 |
Crystal Structure Of A Complex Between Actinomadura R39 Dd-Peptidase And A Boronate Inhibitor
Organism: Actinomadura sp
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-01-26 Classification: HYDROLASE Ligands: EWB, SO4, CO |
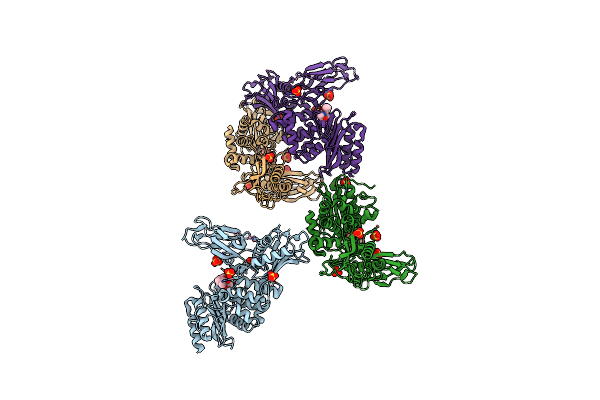 |
Crystal Structure Of A Complex Between Actinomadura R39 Dd-Peptidase And A Boronate Inhibitor
Organism: Actinomadura sp
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-01-26 Classification: HYDROLASE Ligands: EWA, SO4, CO |

