Search Count: 60
 |
Structure Of A De Novo Designed Interleukin-21 Mimetic Complex With Il-21R And Il-2Rg
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp3_R6 Bound To Plpx6 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-03-29 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2023-03-29 Classification: DE NOVO PROTEIN |
 |
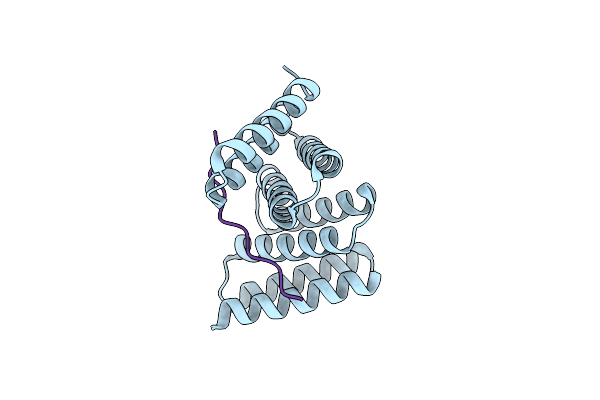
Crystal Structure Of Designed Helical Repeat Protein Rpb_Pew3_R4 Bound To Pawx4 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
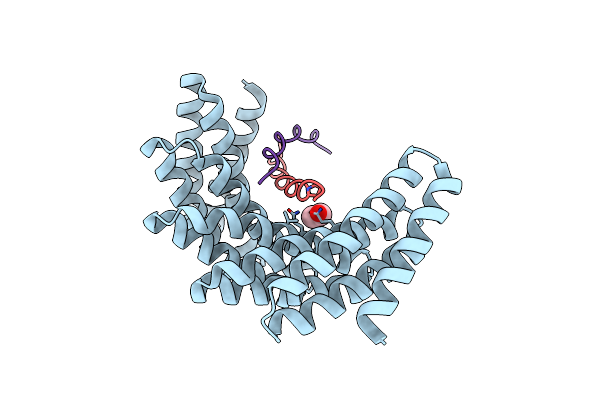
Crystal Structure Of Designed Helical Repeat Protein Rpb_Lrp2_R4 Bound To Lrpx4 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 Bound To Plpx6 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN Ligands: EDO |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 In Alternative Conformation 1 (With Peptide)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 In Alternative Conformation 2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
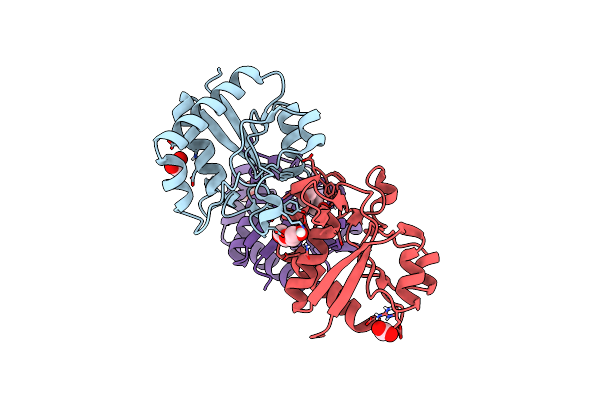
Crystal Structure Of Designed Helical Repeat Protein Rpb_Lrp2_R4 (Proteolysis Fragment?), Forming Pseudopolymeric Filaments
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN Ligands: EDO, PO4 |
 |
Crystal Structure Of Domain A From The Periplasmic Lysine-, Arginine-, Ornithine-Binding Protein (Lao) Of Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-07-14 Classification: PROTEIN BINDING Ligands: ACT, PEG |
 |
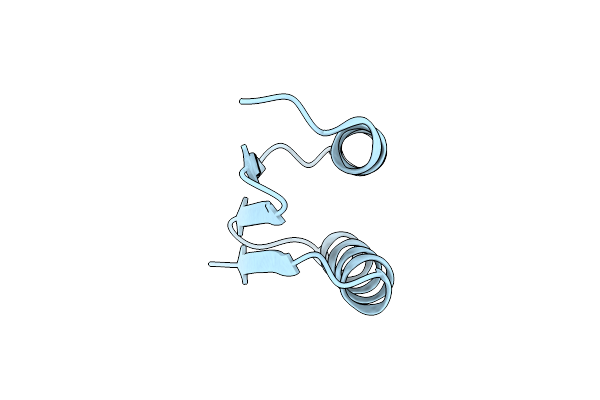
Structure Of The Sars-Cov-2 S 6P Trimer In Complex With The Ace2 Protein Decoy, Ctc-445.2 (State 4)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Structure Of A De Novo Designed Interleukin-2/Interleukin-15 Mimetic Complex With Il-2Rb And Il-2Rg
Organism: Synthetic construct, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN/Protein Binding Ligands: NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-01-10 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-01-10 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-01-10 Classification: DE NOVO PROTEIN |

