Search Count: 16
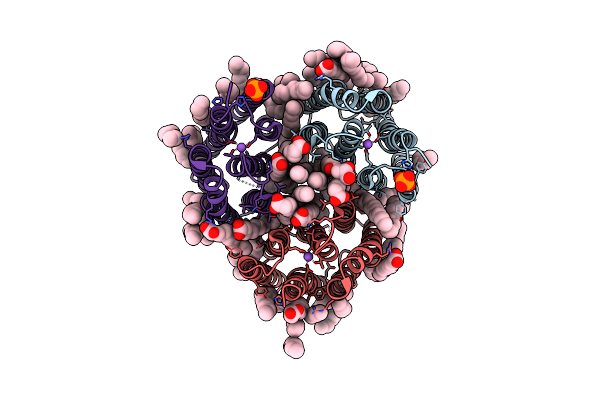 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
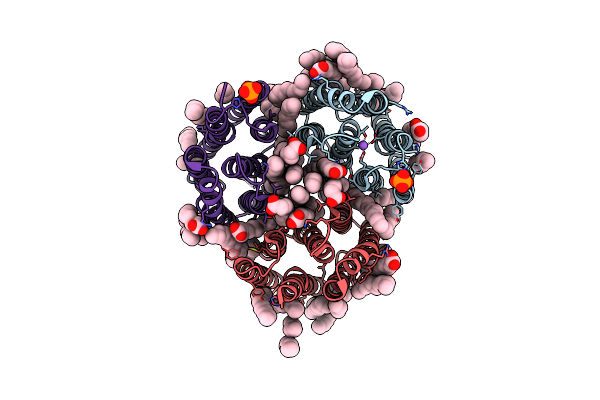 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4, NA |
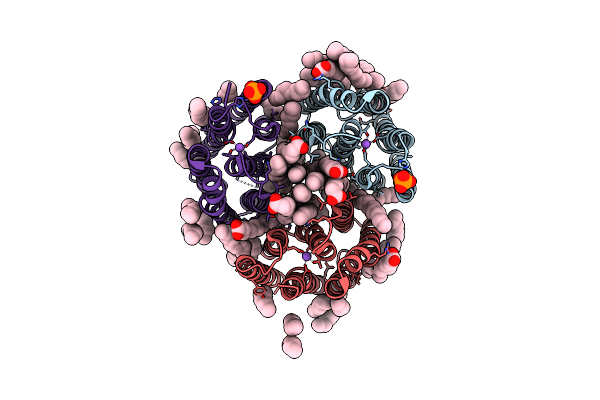 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The L State At Ph 8.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, NA, PO4 |
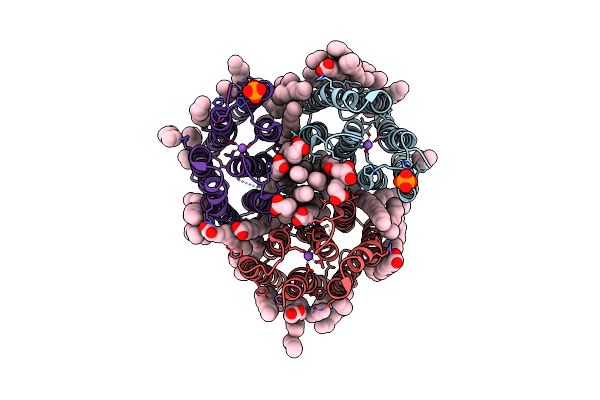 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, NA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 7.0 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: OLA, OLC, LFA, PO4, NA |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 5.2 In The Presence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 7.6 In The Absence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The M State At Ph 7.6 In The Absence Of Sodium At 100K
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 At Room Temperature, 7.5-Ms-Long Snapshots
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Ground State At Ph 8.2 At Room Temperature, 500-Mks-Long Snapshots
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Activated State At Ph 8.2 At Room Temperature, 250-750-Mks-Snapshot
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Light-Driven Inward Proton Pump Xenorhodopsin Bcxer In The Activated State At Ph 8.2 At Room Temperature, 7.5-15-Ms-Snapshot
Organism: Bacillus coahuilensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, PO4 |
 |
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
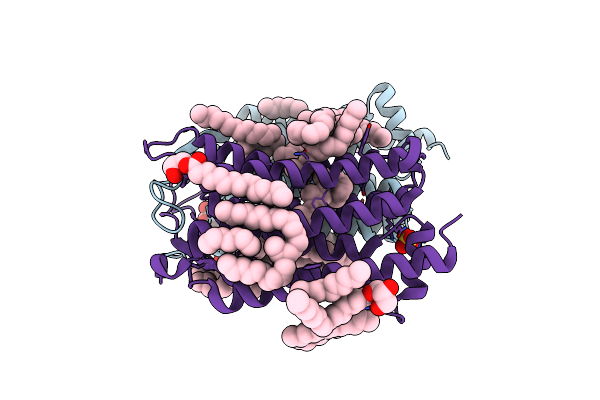 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, SO4, RET, GOL, OLA |
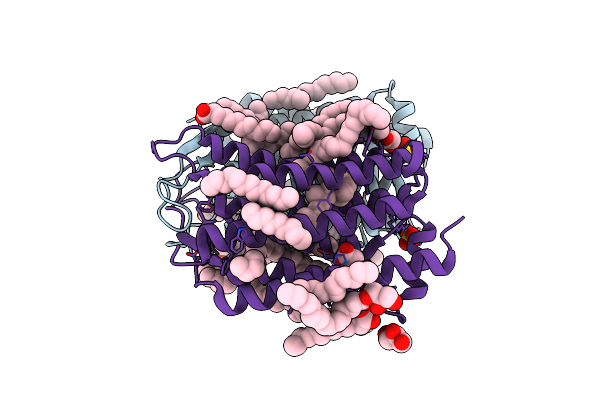 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-11 Classification: MEMBRANE PROTEIN Ligands: ACT, OLA, GOL, OLC, LFA, SO4, RET |

