Search Count: 68
 |
Organism: Syntrophomonas palmitatica jcm 14374, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Bacillus cereus msx-d12
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-02-21 Classification: HYDROLASE |
 |
Organism: Bacillus cereus msx-d12
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: HYDROLASE Ligands: OJC, NAD |
 |
Organism: Bacillus cereus msx-d12
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: HYDROLASE Ligands: RK3, ENA |
 |
Organism: Gordonia otitidis nbrc 100426, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: RNA BINDING PROTEIN |
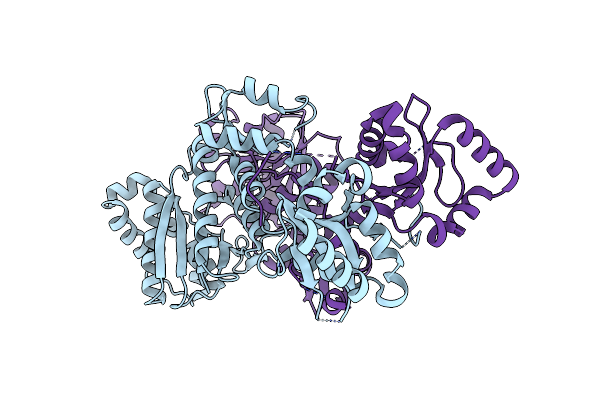 |
Organism: Allochromatium vinosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-13 Classification: HYDROLASE |
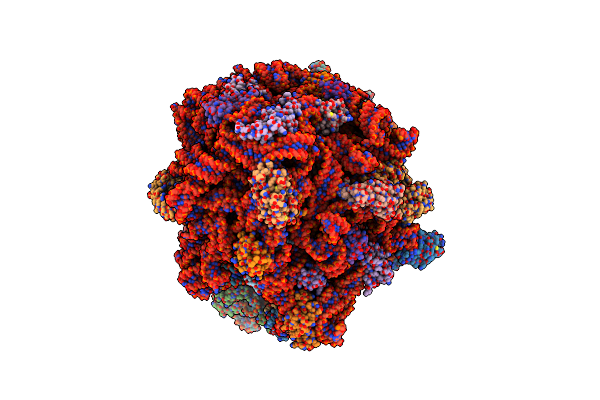 |
Organism: Allochromatium vinosum, Synthetic construct, Streptococcus thermophilus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: RIBOSOME Ligands: ZN, MG |
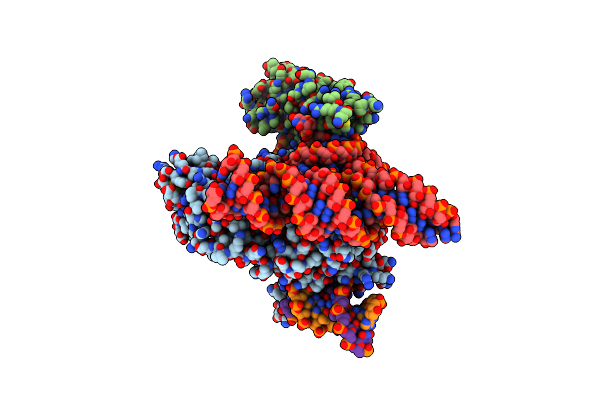 |
Organism: Acidibacillus sulfuroxidans, Acidibacillus sulfooxidans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
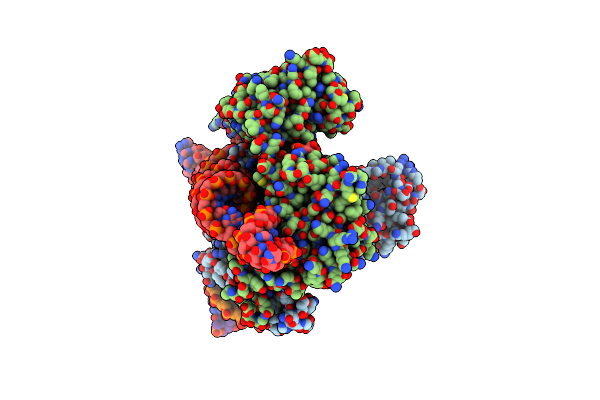 |
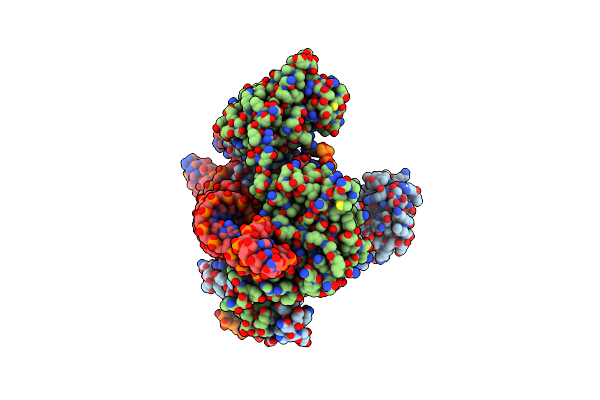
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |
 |
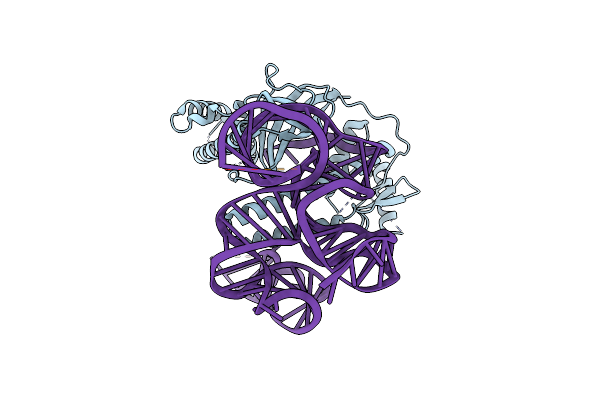
Organism: Sulfoacidibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: ZN, MG |
 |
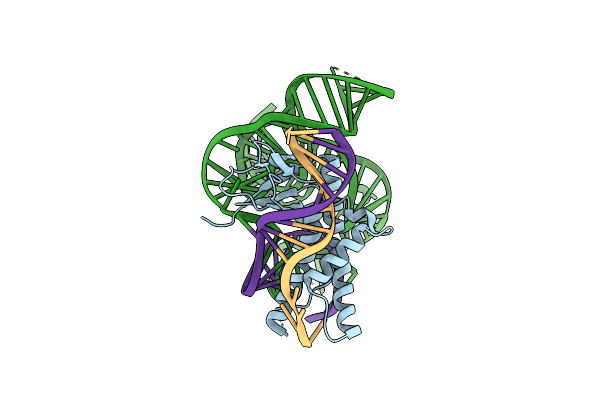
Organism: Deinococcus radiodurans r1
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN |
 |
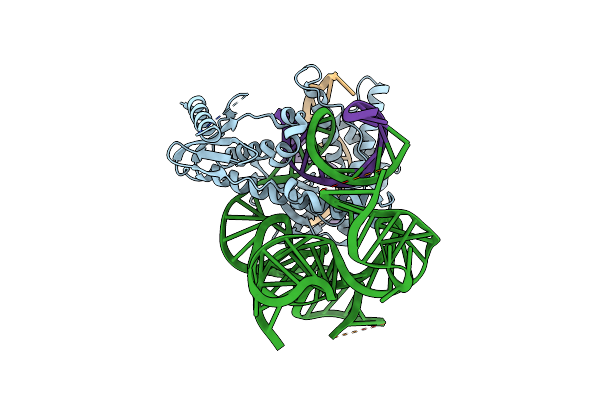
Isdra2 Tnpb In Complex With Rerna And Cognate Dna, Conformation 2 (Ruvc Domain Unresolved)
Organism: Deinococcus radiodurans r1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: RNA BINDING PROTEIN |
 |
Isdra2 Tnpb In Complex With Rerna And Cognate Dna, Conformation 1 (Ruvc Domain Resolved)
Organism: Deinococcus radiodurans r1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Dimeric Porphyromonas Gingivalis Porx, A Type 9 Secretion System Response Regulator.
Organism: Porphyromonas gingivalis (strain atcc baa-308 / w83)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-12-14 Classification: SIGNALING PROTEIN Ligands: BEF, MG, ZN, GOL, FMT |
 |
1.9 Angstrom Crystal Structure Of Dimeric Porx, Co-Crystallized In The Presence Of Zinc
Organism: Porphyromonas gingivalis (strain atcc baa-308 / w83)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-12-14 Classification: SIGNALING PROTEIN Ligands: MG, BEF, ZN, GOL, FMT, CL |
 |
Organism: Porphyromonas gingivalis w83, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2022-12-14 Classification: SIGNALING PROTEIN Ligands: MG, BEF, ZN, GOL, FMT |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: TOXIN/ANTITOXIN |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: AMP, EDO |
 |
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: ACT |
 |
Crystal Structure Of Mono-Ampylated Hepn(R46E) Toxin In Complex With Mnt Antitoxin
Organism: Aphanizomenon flos-aquae 2012/km1/d3
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-12-30 Classification: TOXIN/ANTITOXIN Ligands: 2DA |

