Search Count: 9
 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NDP, 3PE, LMN, ZN, ZMP, PLC, CDL, UQ9, T7X, PSC |
 |
Cryo-Em Structure Of Respiratory Complex I From Yarrowia Lipolytica At 3.2 A Resolution
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, 3PE, NDP, CDL, LMN, PLC, ZN, ZMP, UQ9, T7X, CPL |
 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Resolution:4.04 Å Release Date: 2019-12-11 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NDP, ZN, ZMP |
 |
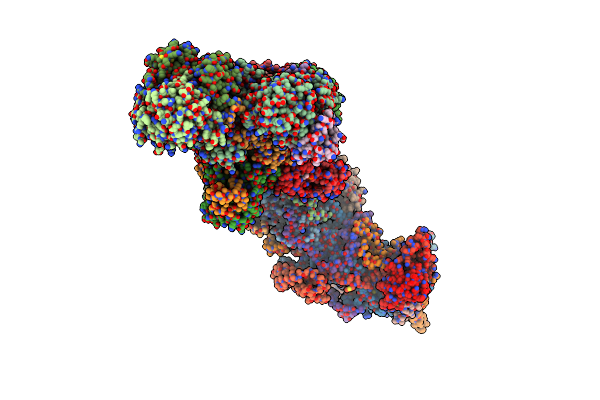
Crystal Structure Of A Variant (Q133C In Psst) Of Yarrowia Lipolytica Complex I
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:3.79 Å Release Date: 2018-12-26 Classification: OXIDOREDUCTASE Ligands: FES, SF4 |
 |
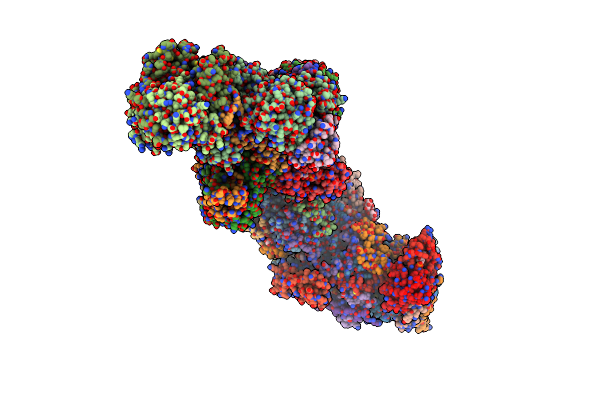
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Resolution:4.32 Å Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NDP, ZN, ZMP, CDL, 3PE |
 |
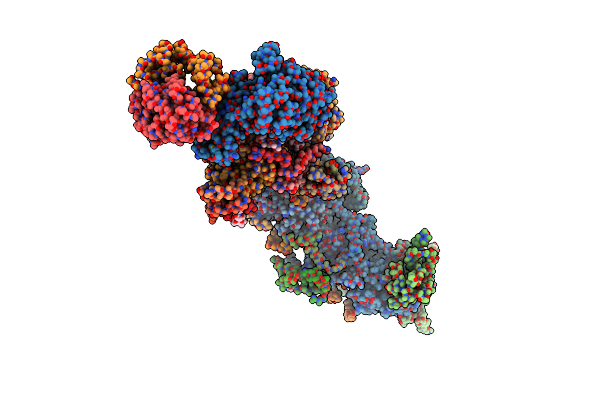
Cryo-Em Structure Of A Respiratory Complex I Assembly Intermediate With Ndufaf2
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: SF4, FES, FMN, NDP, 3PE, LMN, CDL, ZMP, PLC, T7X, CPL |
 |
Crystal Structure Of Mitochondrial Nadh:Ubiquinone Oxidoreductase From Yarrowia Lipolytica.
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-03-25 Classification: OXIDOREDUCTASE Ligands: FES, SF4 |
 |
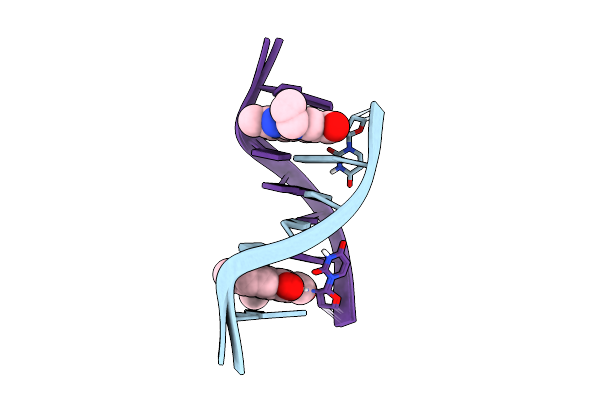
Mini-Haipin Of At Basepairs Having A C12-Alkyl Linker Forming The Loop Region
|
 |
Solution Structure Of The Dna Duplex Acgcgu-Na With A 2' Amido-Linked Nalidixic Acid Residue At The 3' Terminal Nucleotide
|

