Search Count: 9
 |
Crystal Structure Of Fe-S Cluster-Dependent Dehydratase From Paralcaligenes Ureilyticus In Complex With Mg
Organism: Paralcaligenes ureilyticus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-08-30 Classification: LYASE Ligands: FES, MG, BCT, CO2 |
 |
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
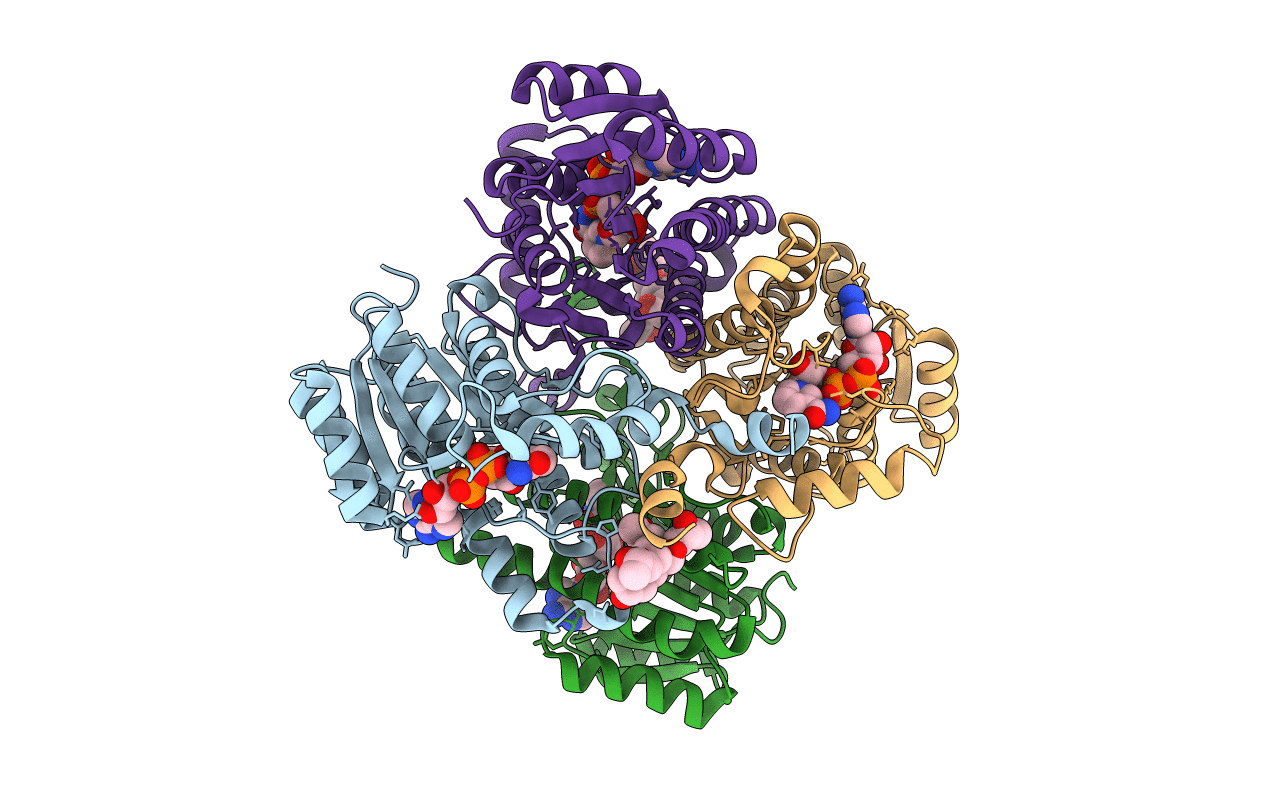 |
Organism: Salvia rosmarinus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: PXN, NAD, CL |
 |
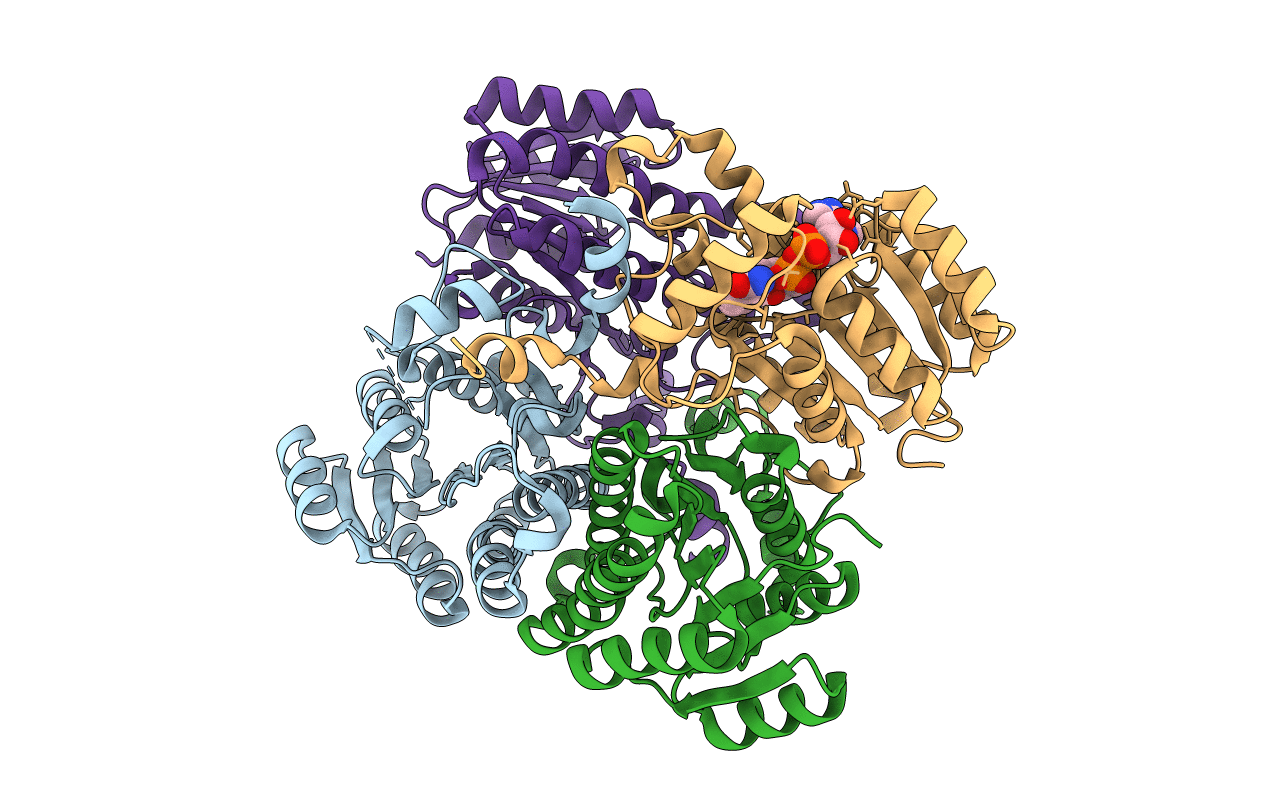
Organism: Salvia rosmarinus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE |
 |
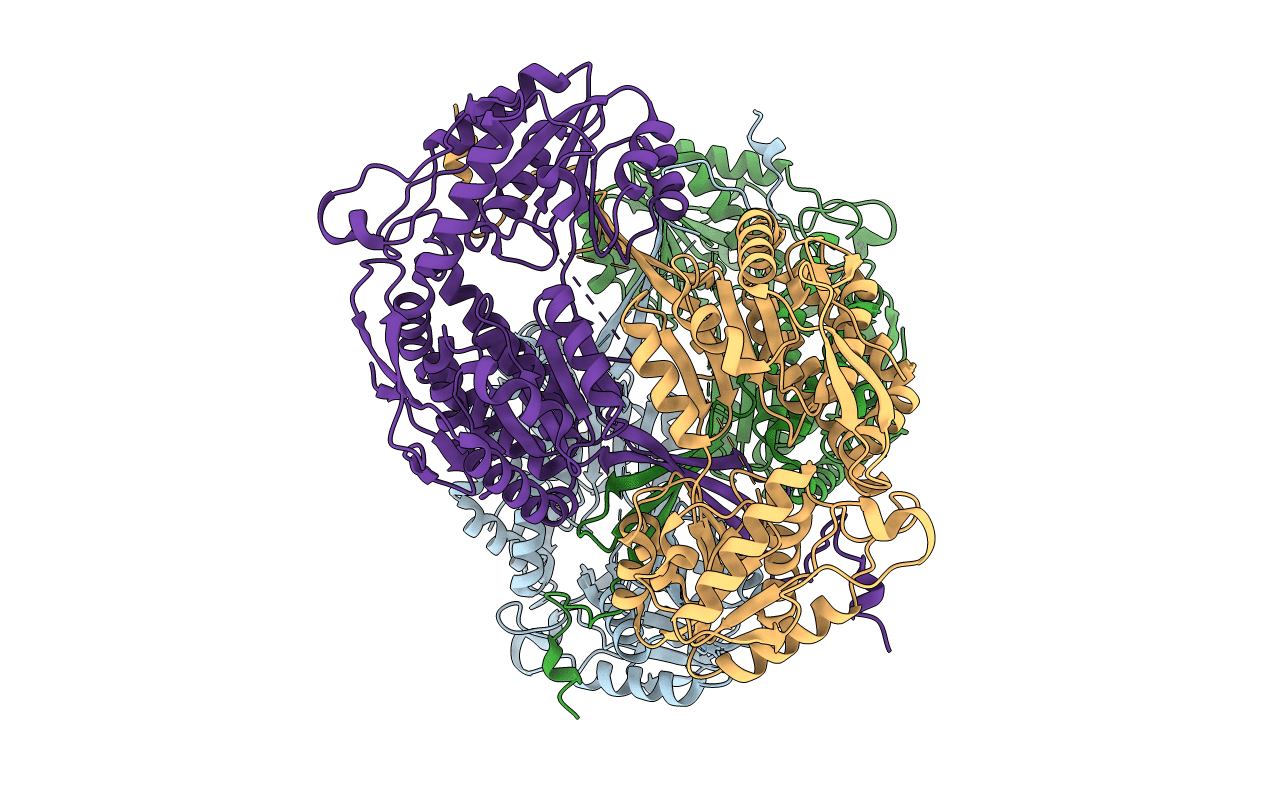
Structure Of The Borneol Dehydrogenases Of Salvia Rosmarinus (High Salt Condition)
Organism: Salvia rosmarinus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-17 Classification: OXIDOREDUCTASE Ligands: CL, NAD |
 |
Mutant Glyceraldehyde Dehydrogenase (F34M+Y399C+S405N) From Thermoplasma Acidophilum
Organism: Thermoplasma acidophilum (strain atcc 25905 / dsm 1728 / jcm 9062 / nbrc 15155 / amrc-c165)
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE |
 |
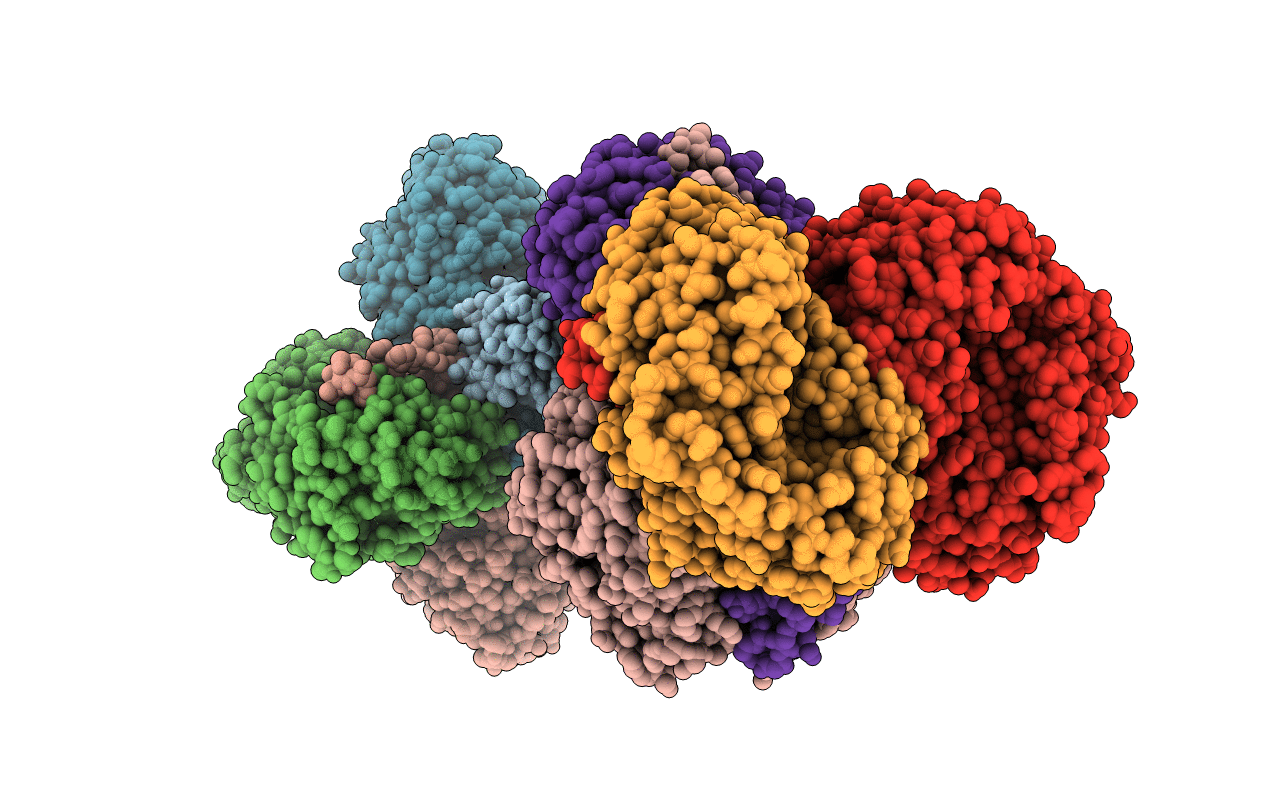
Mutant Glyceraldehyde Dehydrogenase (F34M+S405N) From Thermoplasma Acidophilum
Organism: Thermoplasma acidophilum (strain atcc 25905 / dsm 1728 / jcm 9062 / nbrc 15155 / amrc-c165)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-05-10 Classification: OXIDOREDUCTASE |
 |
Wild-Type Glyceraldehyde Dehydrogenase From Thermoplasma Acidophilum In Complex With Nadp
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-04-05 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Crystal Structure Of Mycobacterium Tuberculosis Ketol-Acid Reductoisomerase In Complex With Mg2+
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2016-02-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, NA |

