Search Count: 46
 |
Crystal Structure Of Thiol Peroxidase From Helicobacter Pylori (Hptx, Reduced)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-05-14 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Thiol Peroxidase Mutant (C94A) In Complex With Metro* (H. Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: A1IAQ, EDO, NA |
 |
Crystal Structure Of Thiol Peroxidase Mutant (C94A) In Complex With Metro-P3* (H. Pylori)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: A1IAR |
 |
Staphylococcus Aureus Clpp In Complex With The Natural Product Beta-Lactone Inhibitor Cystargolide A At 2.0 A Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: VSZ, EDO |
 |
Structure Of Staphylococcus Aureus Clpp Bound To The Covalent Active Site Inhibitor Cystargolide A
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: VSZ |
 |
Photorhabdus Lamondii Clpp In Complex With The Natural Product Beta-Lactone Inhibitor Cystargolide A At 2.5 A Resolution
Organism: Photorhabdus laumondii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: VSZ |
 |
Staphylococcus Aureus Clpp In Complex With The Natural Product Beta-Lactone Inhibitor Cystargolide A At 2.7 A Resolution
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: VSZ |
 |
Structure Of Yeast 20S Proteasome In Complex With The Natural Product Beta-Lactone Inhibitor Cystargolide A
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: MG, CL, VSZ |
 |
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-11-15 Classification: LIGASE |
 |
Crystal Structure Of Lpla1 In Complex With Lipoic Acid (Listeria Monocytogenes)
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-06-07 Classification: LIGASE Ligands: LPA, CL, SO4, EDO |
 |
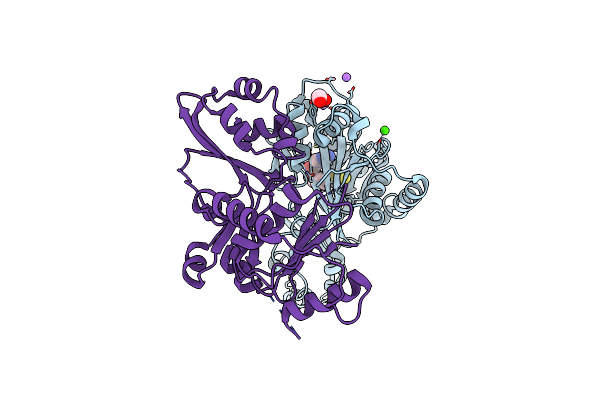
Crystal Structure Of Lpla1 In Complex With Lipoyl-Amp (Listeria Monocytogenes)
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-06-07 Classification: LIGASE Ligands: LAQ, MG, SO4, EDO, GOL |
 |
Crystal Structure Of Lpla1 In Complex With The Inhibitor C3 (Listeria Monocytogenes)
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-06-07 Classification: LIGASE Ligands: VK9, CA, NA, EDO |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-09-21 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-26 Classification: TRANSLATION Ligands: ACT, EDO, NA |
 |
Cryo-Em Structure Of The Clpx Component Of The Clpxp1/2 Degradation Machinery.
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2019-10-16 Classification: TRANSPORT PROTEIN |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2019-10-16 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The Hclpp Y118A Mutant With An Activating Small Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: FJT, EDO |
 |
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL |
 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With A Pyridoxal 5' Phosphate-Derivative
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EOW, NA, MPD, ACT, CL, MRD, GOL |
 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With An Pyridoxal-6- Phosphate Derivative
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EM2, SO4, NA, CL |

