Search Count: 17
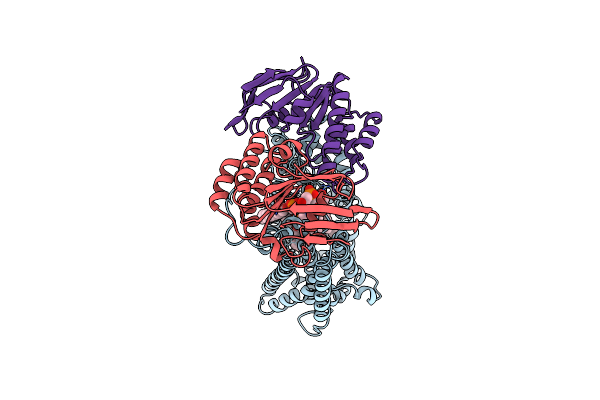 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: L9Q, CDL |
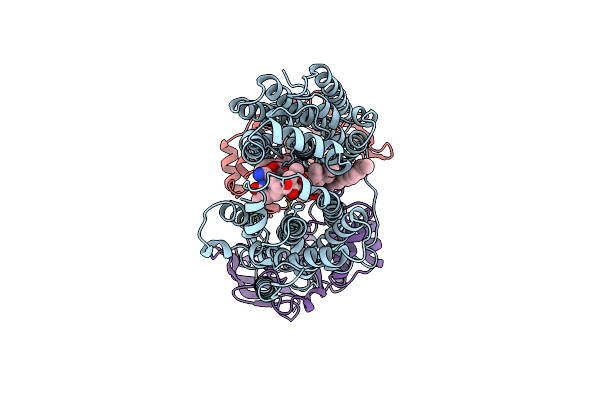 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: RFP, CDL |
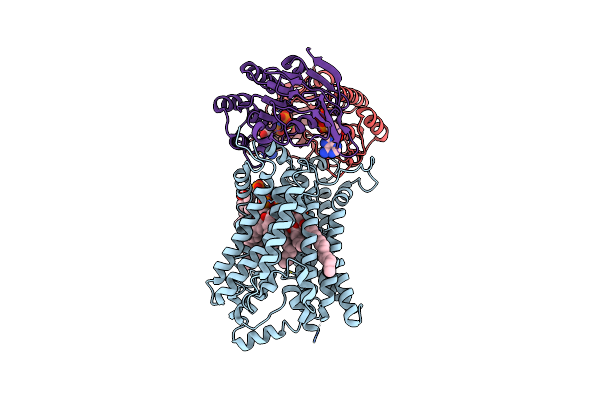 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: CDL, ANP, MG |
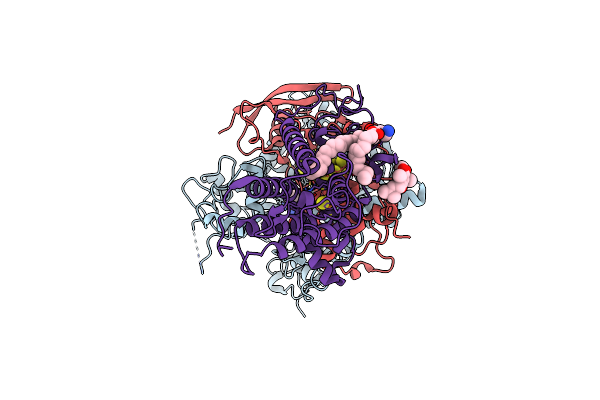 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: OXIDOREDUCTASE Ligands: FAD, FES, SF4, F3S, UQ1, PEV |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: OXIDOREDUCTASE Ligands: FAD, FES, SF4, F3S, PEV |
 |
Organism: Mycolicibacterium smegmatis mc2 51
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2020-05-27 Classification: OXIDOREDUCTASE Ligands: PEV, HEM, MQ9, CDL, LPP, PIE, FAD, FES, SF4, F3S |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: BR, CA |
 |
Structural Basis For Activity Of Tric Counter-Ion Channels In Calcium Release
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CA, CL |
 |
Structural Basis For Activity Of Tric Counter-Ion Channels In Calcium Release
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:3.79 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Structural Basis For Activity Of Tric Counter-Ion Channels In Calcium Release
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-01 Classification: MEMBRANE PROTEIN Ligands: CA, CL, BQ9 |
 |
Organism: Mycobacterium smegmatis mc2 51
Method: ELECTRON MICROSCOPY Release Date: 2018-11-14 Classification: ELECTRON TRANSPORT Ligands: CU, HEA, CDL, 9Y0, PLM, 9XX, 9YF, HEM, MQ9, HEC, FES |

