Search Count: 94
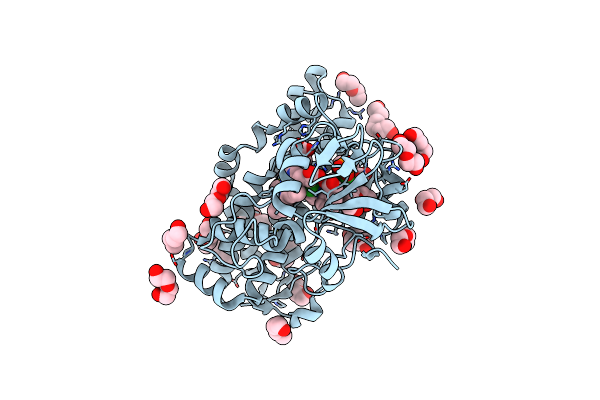 |
The Crystal Structure Of Cytochrome P450 105D7 From Streptomyces Avermitilis In Complex With Diclofenac
Organism: Streptomyces avermitilis ma-4680 = nbrc 14893
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-11-05 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, PGE, PEG, PO4 |
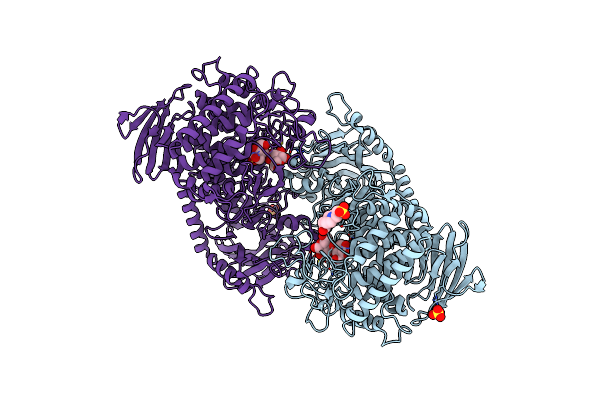 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Sulfate And Isofagomine
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, SO4, IFM, EPE |
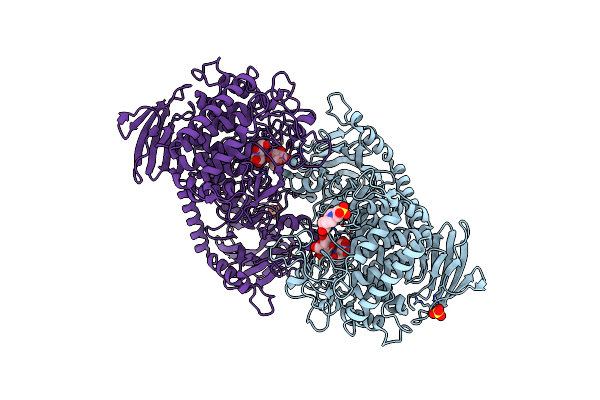 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Sulfate And 1-Deoxynojirimycin
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, SO4, NOJ, EPE |
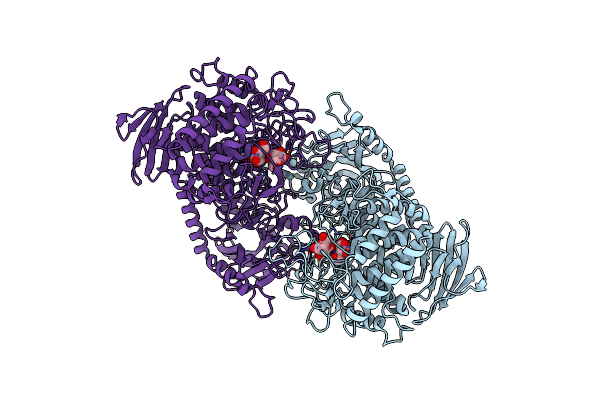 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Complexed With Phosphate And 1-Deoxynojirimycin
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-09-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: BGC, PO4, NOJ |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Triple Mutant
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-03-23 Classification: TRANSFERASE Ligands: BGC, PO4, K |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase W488F Mutant
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2010-12-22 Classification: TRANSFERASE Ligands: BGC, SO4 |
 |
Crystal Structure Of Cellvibrio Gilvus Cellobiose Phosphorylase Histidine Mutant
Organism: Cellvibrio gilvus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-12-22 Classification: TRANSFERASE Ligands: BGC, PO4, K, GOL |
 |
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, TPP, NA, EDO, 2PE |
 |
Phosphoketolase From Bifidobacterium Breve Complexed With 2-Acetyl-Thiamine Diphosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, HTL, NA, EDO |
 |
Phosphoketolase From Bifidobacterium Breve Complexed With Dihydroxyethyl Thiamine Diphosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, THD, NA, EDO, 2PE |
 |
Phosphoketolase From Bifidobacterium Breve Complexed With Inorganic Phosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-08-25 Classification: LYASE Ligands: TPP, MG, PO4, GOL |
 |
H64A Mutant Of Phosphoketolase From Bifidobacterium Breve Complexed With A Tricyclic Ring Form Of Thiamine Diphosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, HTD, NA, EDO |
 |
H142A Mutant Of Phosphoketolase From Bifidobacterium Breve Complexed With Acetyl Thiamine Diphosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, HTL, NA, EDO, PG4 |
 |
H320A Mutant Of Phosphoketolase From Bifidobacterium Breve Complexed With Acetyl Thiamine Diphosphate
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, HTL, NA, EDO |
 |
Organism: Bifidobacterium breve
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: LYASE Ligands: MG, TPP, EDO |
 |
Organism: Streptomyces avermitilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-04-07 Classification: OXIDOREDUCTASE/ANTIBIOTIC Ligands: HEM, FLI, SO4 |
 |
Organism: Streptomyces avermitilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-07 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In Complex With Cellopentaose
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-31 Classification: HYDROLASE Ligands: CA, PO4 |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In A Ligand-Free Form
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Crystal Structure Of Carbohydrate-Binding Module Family 28 From Clostridium Josui Cel5A In Complex With Cellobiose
Organism: Clostridium josui
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CA, PO4, GOL |

