Search Count: 16
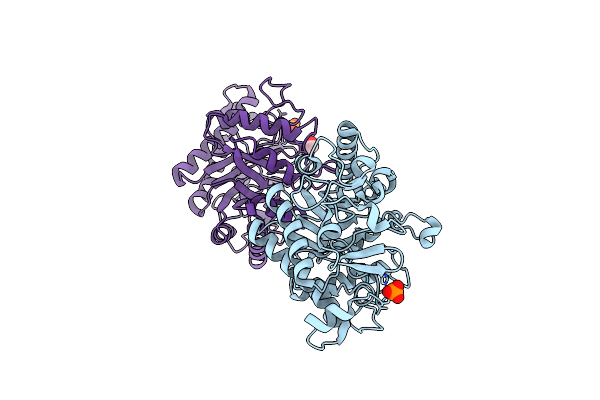 |
Se-Sad Structure Of The Functional Region Of Cwp19 From Clostridium Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-01 Classification: HYDROLASE Ligands: PO4, EDO |
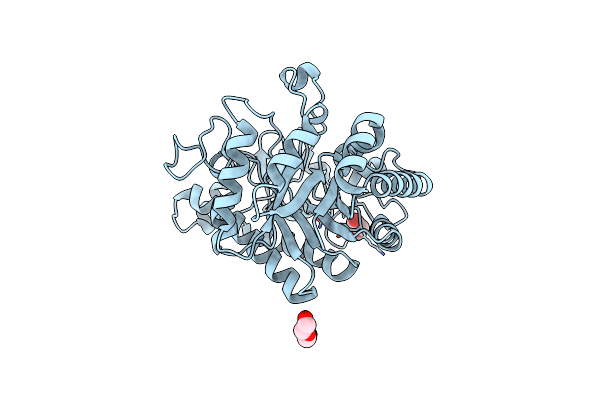 |
High Resolution Structure Of The Functional Region Of Cwp19 From Clostridium Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-11-01 Classification: HYDROLASE Ligands: CL, EDO, PEG |
 |
Organism: Clostridioides difficile r20291
Method: X-RAY DIFFRACTION Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: SO4 |
 |
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-04-01 Classification: HYDROLASE Ligands: PEG, 1PE |
 |
Organism: Peptoclostridium difficile qcd-32g58
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, JEF |
 |
Organism: Peptoclostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, PGE, GOL |
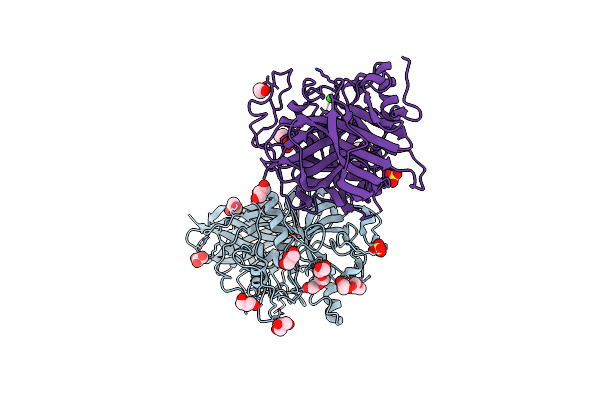 |
The Crystal Structure Of The Cysteine Protease And Lectin-Like Domains Of Cwp84, A Surface Layer Associated Protein Of Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-05-14 Classification: HYDROLASE Ligands: SO4, GOL, CA, PGE |
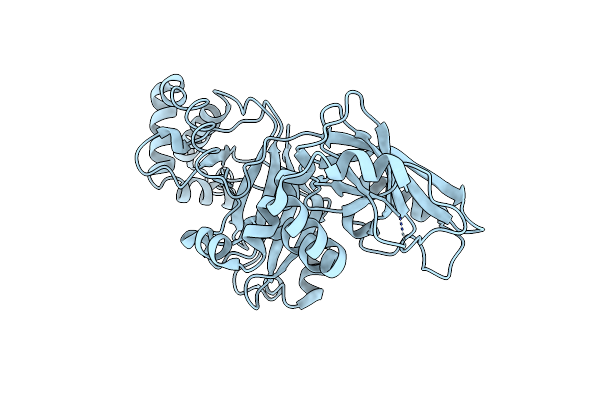 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-18 Classification: TRANSFERASE |
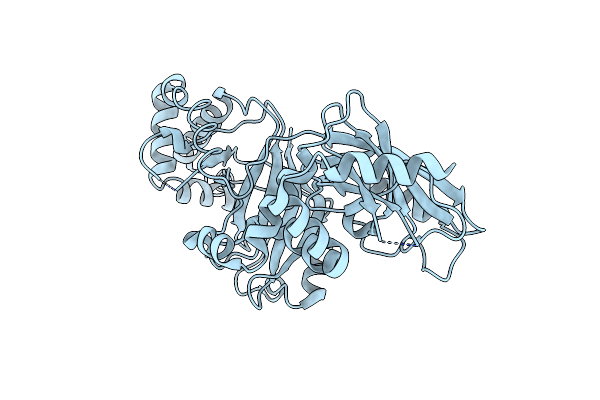 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-08-18 Classification: TRANSFERASE |
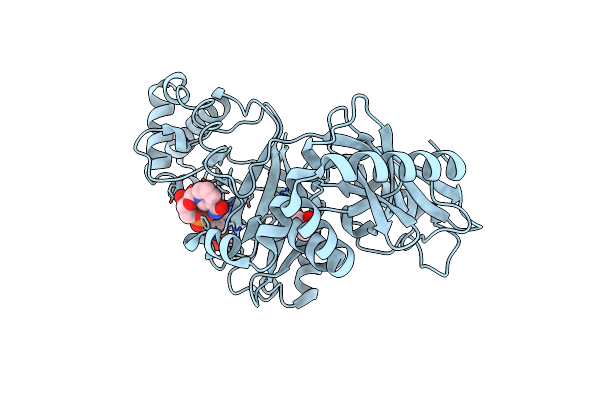 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2009-08-18 Classification: TRANSFERASE Ligands: NDP, GOL |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-08-18 Classification: TRANSFERASE Ligands: NAD, GOL |
 |
Structural Basis For Substrate Recognition In The Enzymatic Component Of Adp-Ribosyltransferase Toxin Cdta From Clostridium Difficile
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-18 Classification: TRANSFERASE |
 |
Molecular Recognition Of An Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme By Rala Gtpase
Organism: Homo sapiens, Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2005-04-15 Classification: TRANSFERASE Ligands: GDP, MG |
 |
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2004-07-29 Classification: TRANSFERASE Ligands: V4O, VO4, GOL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2003-08-28 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2003-08-28 Classification: TRANSFERASE Ligands: NAD |

