Search Count: 7
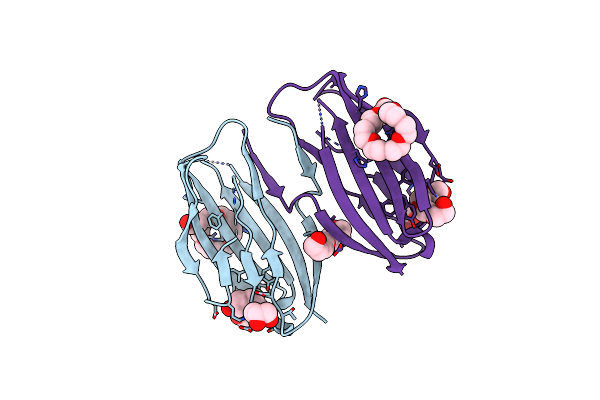 |
Parallel Homodimer Structures Of Voltage-Gated Sodium Channel Beta4 For Cell-Cell Adhesion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: P33, PG4, GOL |
 |
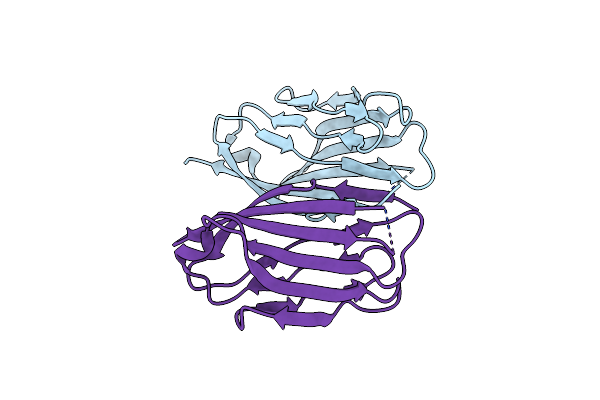
Parallel Homodimer Structures Of The Extracellular Domains Of The Voltage-Gated Sodium Channel Beta4 Subunit Explain Its Role In Cell-Cell Adhesion
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: GOL |
 |
Structure-Based Site-Directed Photo-Crosslinking Analyses Of Multimeric Cell-Adhesive Interactions Of Vgsc Beta Subunits
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-08 Classification: METAL TRANSPORT |
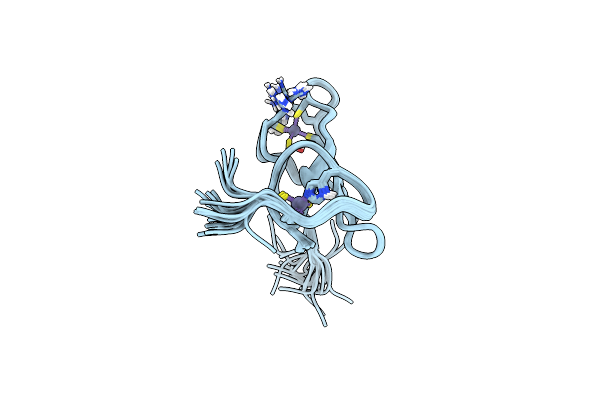 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2014-07-16 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
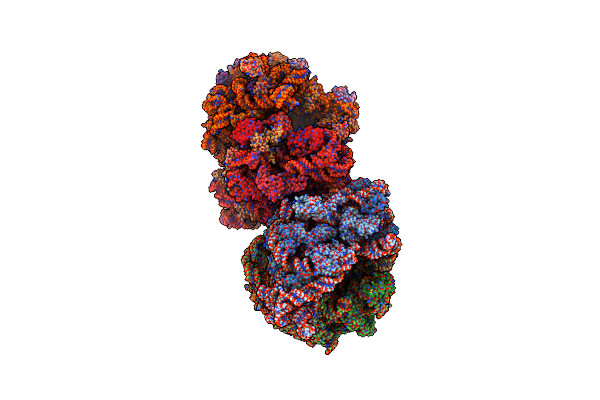
Crystal Structure Of The Bacterial Ribosome From Escherichia Coli In Complex With Spectinomycin.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.93 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, SCM, ZN |
 |
Crystal Structure Of The Bacterial Ribosome From Escherichia Coli In Complex With Spectinomycin And Neomycin.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: NMY, MG, SCM, ZN |
 |
Crystal Structure Of The Bacterial Ribosome From Escherichia Coli In Complex With Hygromycin B.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, HYG, ZN |

