Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PEPTIDE NUCLEIC ACID |
 |
Synchrotron X-Ray Crystal Structure Of Oxygen-Bound F87A/F393H P450Bm3 With Decoy C7Prophe (N-Enanthyl-L-Prolyl-L-Phenylalanine) And Substrate Styrene At 2 Mgy X-Ray Dose
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, SYN, OXY, D0L, GOL |
 |
Xfel Crystal Structure Of The Oxidized Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Oxidized Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Reduced Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Reduced Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Oxygen-Bound Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, OXY, GOL |
 |
Xfel Crystal Structure Of The Oxygen-Bound Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, OXY, GOL |
 |
Crystal Structure Of Cytochrome P450Bm3 Iii-10C1 Mutant Heme Domain With N-Decanoyl-L-Homoserine Lactone
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, HL0, DMS, GOL |
 |
Crystal Structure Of Cytochrome P450Bm3 Vi-18A12 Mutant Heme Domain With N-Decanoyl-L-Homoserine Lactone
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, HL0, DMS, GOL |
 |
Crystal Structure Of Cytochrome P450Bm3 V-19A14 Mutant Heme Domain With N-Decanoyl-L-Homoserine Lactone
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, HL0, DMS, GOL |
 |
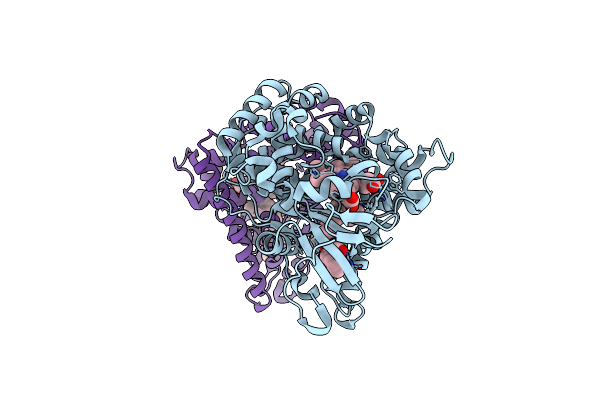
Crystal Structure Of The Hemophore Hasa From Pseudomonas Protegens Pf-5 Capturing Fe-Tetraphenylporphyrin
Organism: Pseudomonas protegens pf-5
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-03-27 Classification: TRANSPORT PROTEIN Ligands: MQP, NHE, GOL, NA |
 |
Crystal Structure Of The Holo Form Of The Hemophore Hasa From Pseudomonas Protegens Pf-5
Organism: Pseudomonas protegens pf-5
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-27 Classification: TRANSPORT PROTEIN Ligands: HEM, GOL, PO4 |
 |
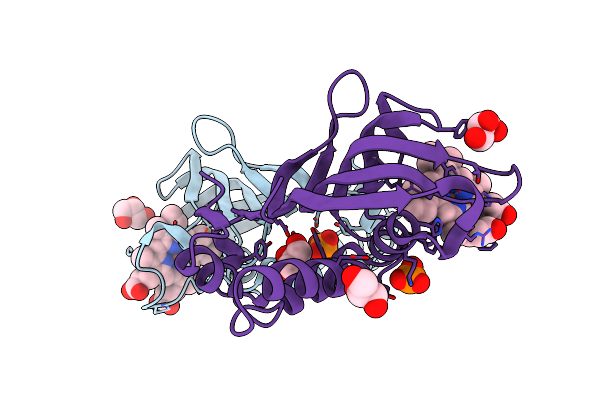
Crystal Structure Of P450Bm3 With N-(3-Cyclohexylpropanoyl)-L-Pipecolyl-L-Phenylalanine
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: HEM, I8F |
 |
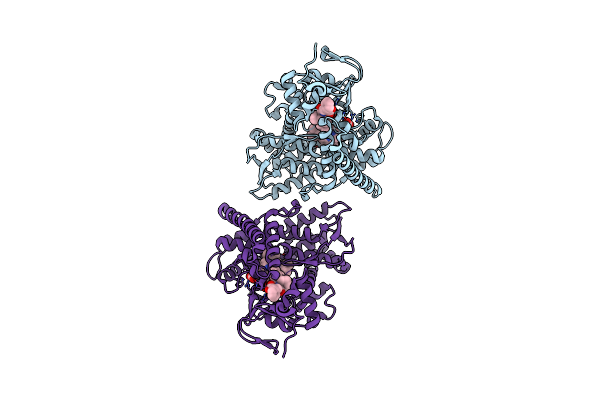
Crystal Structure Of The Cyp102A5 Haem Domain Isolated From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: HEM, PLM |
 |
Structure Of The Oxomolybdenum Mesoporphyrin Ix-Reconstituted Cyp102A1 Haem Domain With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styerene
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: MI9, D0L, SYN, SOR |
 |
Structure Of The Oxomolybdenum Mesoporphyrin Ix-Reconstituted Cyp102A1 F87A Mutant Haem Domain With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: MI9, D0L, SYN, GOL, CL |
 |
Structure Of The Oxomolybdenum Mesoporphyrin Ix-Reconstituted Cyp102A1 F87V Mutant Haem Domain With N-(5-Cyclohexyl)Valeroyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: MI9, GKX, SYN, GOL, CL |
 |
Structure Of The Cyp102A1 F87A Haem Domain With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, CL, GOL |
 |
Crystal Structure Of The Cyp102A1 (P450Bm3) Heme Domain With N-Hexadecanoyl-L-Homoserine
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: HEM, 8PD, GOL |