Search Count: 36
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 100 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Dark State Sfx Structure Of Hen Egg White Lysozyme Microcrystals Immobilized With A Mn Carbonyl Complex At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 10 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
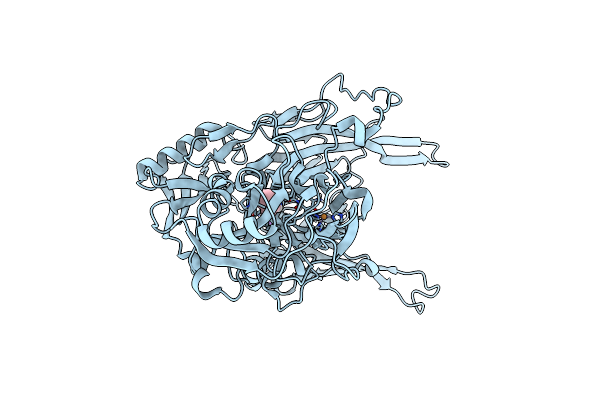
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation With 40 Microjoules Laser Intensity At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Globiformis Anaerobically Reduced By Phenylethylamine At Pd 9.0
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.09 Å, 1.67 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
 |
Holo Form Of N381A Mutant Of Copper Amine Oxidase From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
 |
Catalytic Intermediate Structure Of N381A Mutant Of Copper Amine Oxidase From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: CU, HY1, GOL |
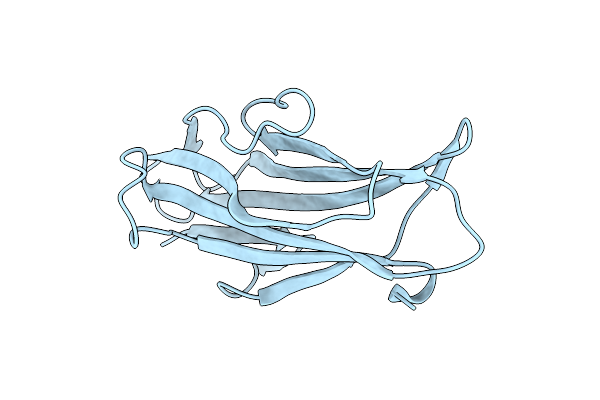 |
Organism: Porphyromonas gingivalis atcc 33277
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-22 Classification: SIGNALING PROTEIN |
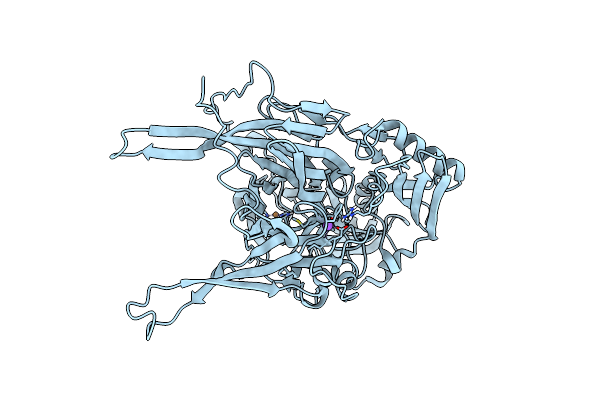 |
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.1400 Å, 1.7200 Å Release Date: 2020-04-29 Classification: OXIDOREDUCTASE Ligands: CU, NA |
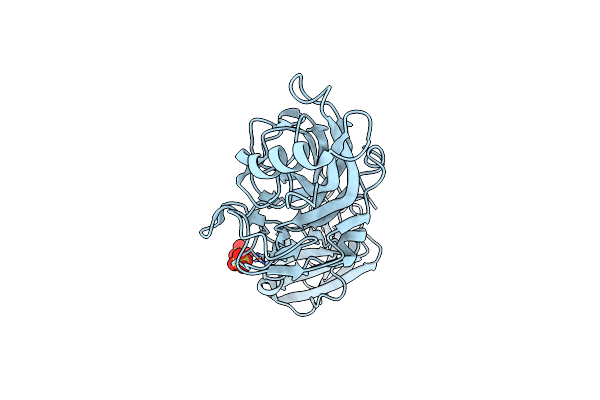 |
Organism: Porphyromonas gingivalis tdc60
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: CELL ADHESION Ligands: SO4 |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-15 Classification: CELL ADHESION Ligands: GOL, SO4, SRT |
 |
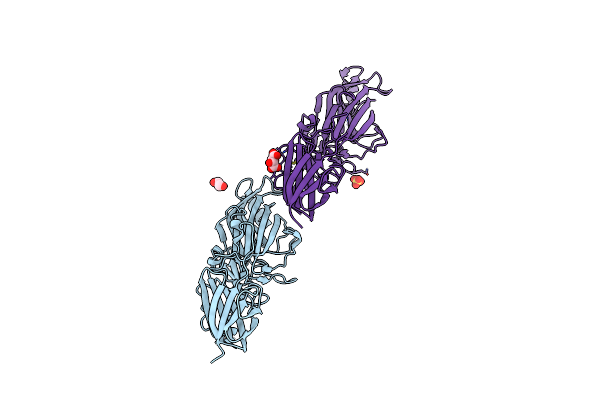
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: CELL ADHESION |
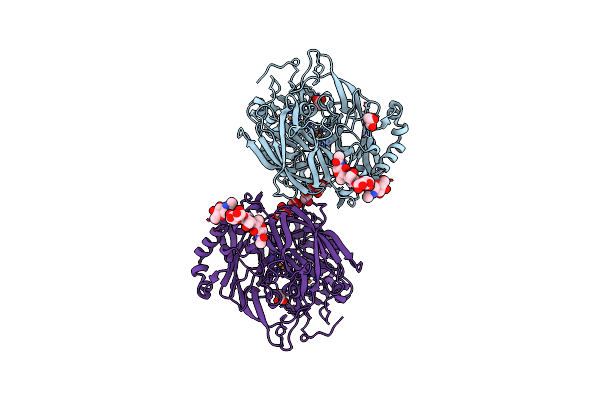 |
High Resolution Structure Of Bilirubin Oxidase From Myrothecium Verrucaria - M467Q Mutant, Aerobically Prepared
Organism: Myrothecium verrucaria
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2018-11-21 Classification: OXIDOREDUCTASE Ligands: CU, NAG, GOL, ACT |
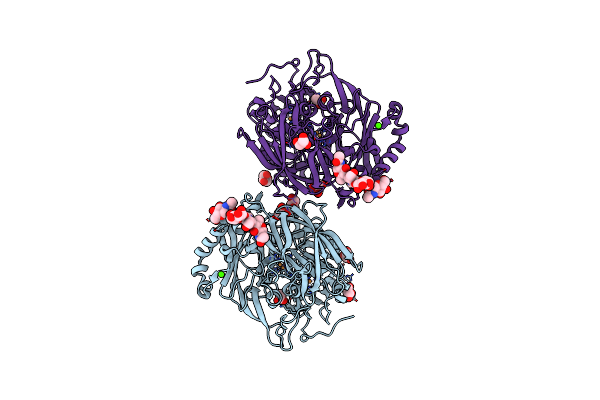 |
High Resolution Structure Of Bilirubin Oxidase From Myrothecium Verrucaria - M467Q Mutant, Anaerobically Prepared
Organism: Myrothecium verrucaria
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-11-21 Classification: OXIDOREDUCTASE Ligands: CU, NAG, ACT, GOL, CA |
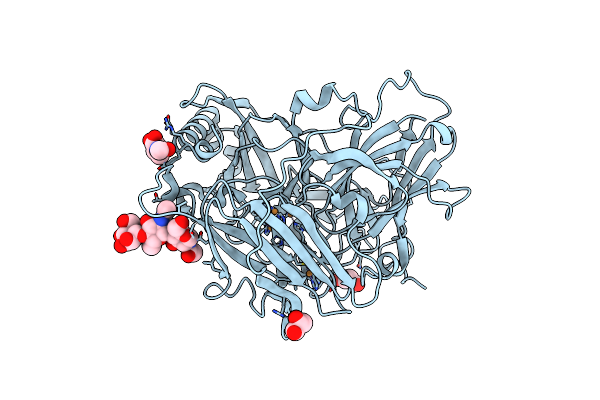 |
High Resolution Structure Of Bilirubin Oxidase From Myrothecium Verrucaria - Wild Type
Organism: Myrothecium verrucaria
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2018-11-21 Classification: OXIDOREDUCTASE Ligands: CU, NAG, GOL |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-07-25 Classification: ELECTRON TRANSPORT |
 |
Crystal Structure Of A Putative Adhesin (Bacova_02677) From Bacteroides Ovatus Atcc 8483 At 3.00 A Resolution (Psi Community Target, Nakayama)
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-10-14 Classification: CELL ADHESION Ligands: SO4 |
 |
Crystal Structure Of An Immunoreactive 32 Kda Antigen Pg49 (Pg_0181) From Porphyromonas Gingivalis W83 At 1.45 A Resolution (Psi Community Target, Nakayama)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-12-03 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: EDO, ACN |
 |
Crystal Structure Of A Fimbrial Protein (Bvu_2522) From Bacteroides Vulgatus Atcc 8482 At 2.55 A Resolution
Organism: Bacteroides vulgatus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-07-02 Classification: CELL ADHESION Ligands: SO4, CL |

