Search Count: 15
 |
Sars Cov-2 Spike Protein, Bristol Uk Deletion Variant, Closed Conformation, C3 Symmetry
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
Sars Cov-2 Spike Protein, Bristol Uk Deletion Variant, Closed Conformation, C1 Symmetry
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
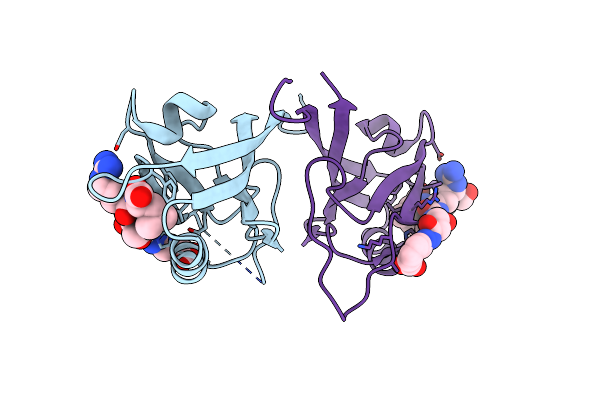
Crystal Structure Of Shank1 Pdz In Complex With L6F Mutant Of The C-Terminal Hexapeptide From Gkap
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-07-07 Classification: PEPTIDE BINDING PROTEIN |
 |
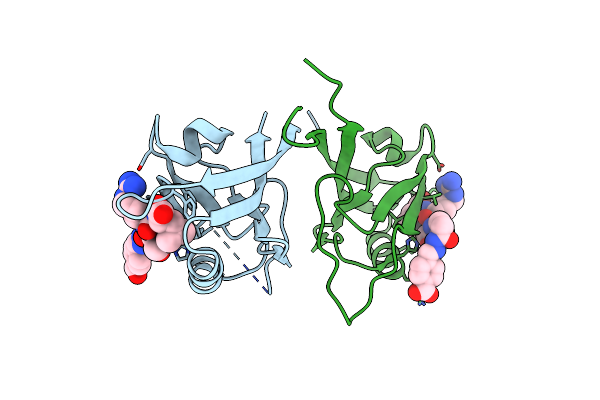
Crystal Structure Of Shank1 Pdz In Complex With A Peptide-Small Molecule Hybrid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-01-20 Classification: PEPTIDE BINDING PROTEIN Ligands: PEG, PWN, THR, ARG, LEU |
 |
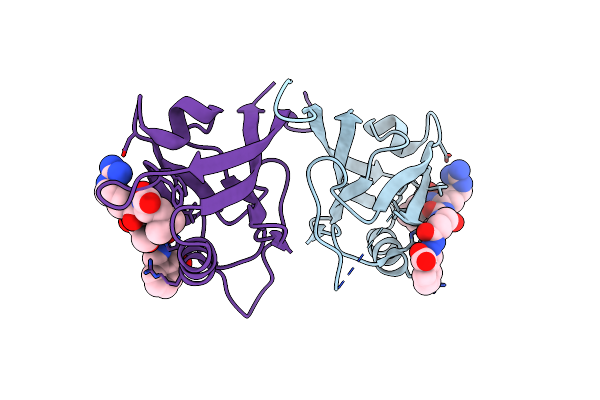
Crystal Structure Of Shank1 Pdz In Complex With A Peptide-Small Molecule Hybrid
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-01-20 Classification: PEPTIDE BINDING PROTEIN Ligands: PWQ |
 |
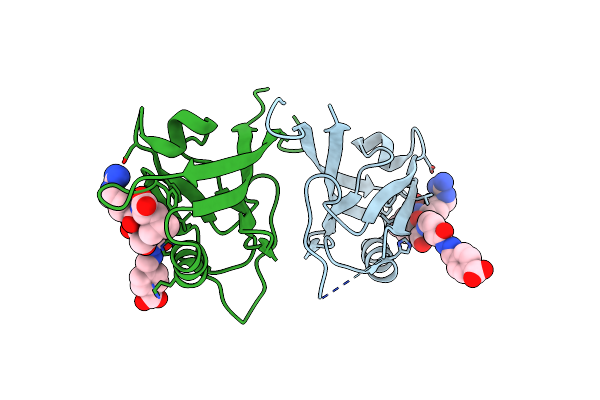
Crystal Structure Of Shank1 Pdz In Complex With A Peptide-Small Molecule Hybrid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-20 Classification: PEPTIDE BINDING PROTEIN Ligands: PWT, THR, ARG, LEU |
 |
Crystal Structure Of Shank1 Pdz In Complex With A Peptide-Small Molecule Hybrid
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2021-01-20 Classification: PEPTIDE BINDING PROTEIN Ligands: PWW |
 |
Crystal Structure Of Neuropilin-1 B1 Domain In Complex With Sars-Cov-2 S1 C-End Rule (Cendr) Peptide
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: PEG, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-09-30 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-09-30 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2015-05-27 Classification: OXIDOREDUCTASE |
 |
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-05-27 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
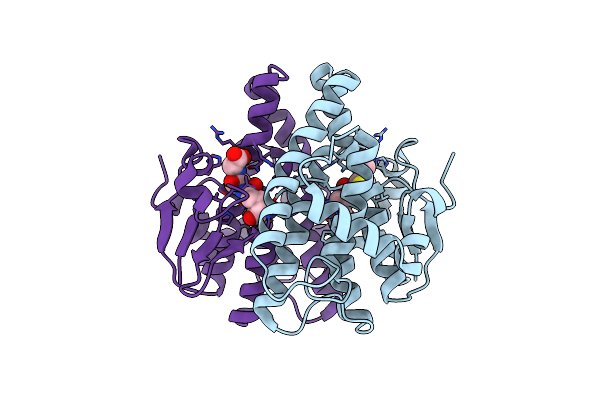 |
Holo Structure Of Maleyl Pyruvate Isomerase, A Bacterial Glutathione- S-Transferase In Zeta Class
Organism: Ralstonia sp. u2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-09-16 Classification: ISOMERASE Ligands: GSH |
 |
Structure Of Maleyl Pyruvate Isomerase, A Bacterial Glutathione-S- Transferase In Zeta Class, In Complex With Substrate Analogue Dicarboxyethyl Glutathione
Organism: Ralstonia sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2008-07-22 Classification: ISOMERASE Ligands: TGG, NA, ACT |

