Search Count: 73
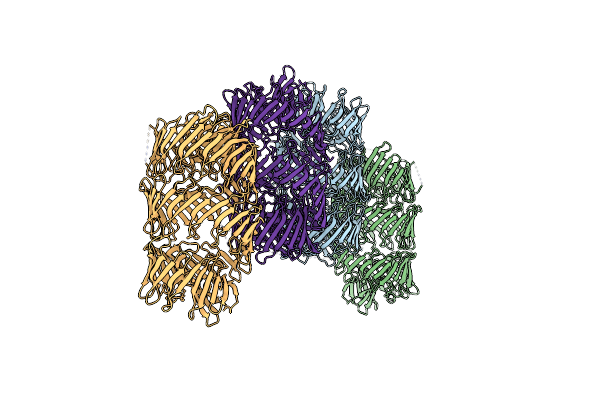 |
T6Ss-Linked Rhs Repeat Protein - Advenella Mimigardefordensis Vgrg-Rhs Core
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2024-09-25 Classification: CELL INVASION |
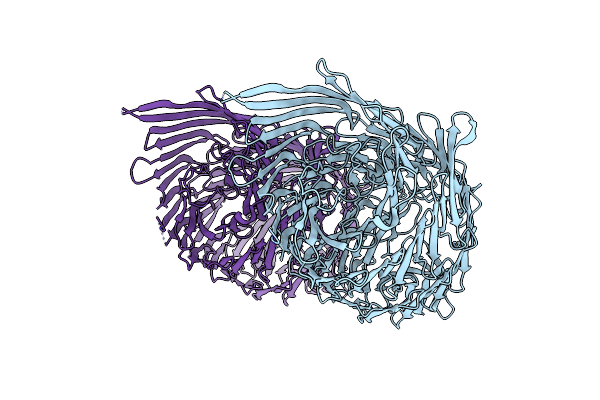 |
T6Ss-Linked Rhs Repeat Protein - Salmonella Bongori Rhs-Core Domain Extended N-Terminus
Organism: Salmonella bongori n268-08
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2024-09-25 Classification: CELL INVASION |
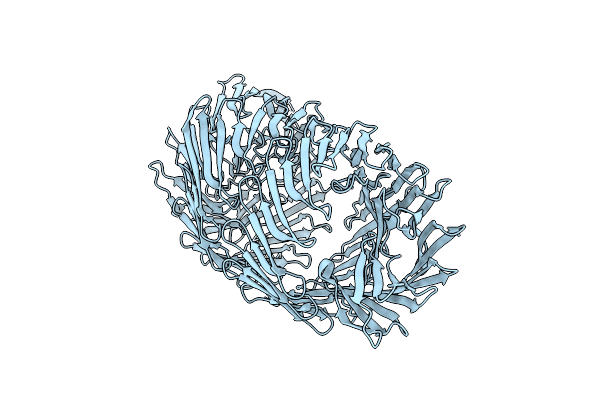 |
T6Ss-Linked Rhs Repeat Protein - Salmonella Bongori Rhs-Core Domain With Toxin Domain
Organism: Salmonella bongori n268-08
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-09-25 Classification: CELL INVASION |
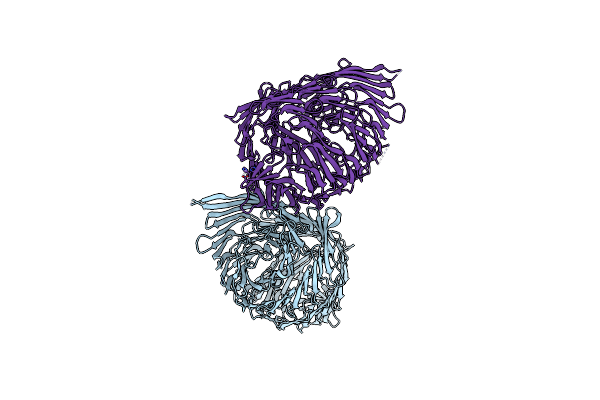 |
Organism: Salmonella bongori n268-08
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-25 Classification: CELL INVASION Ligands: ZN |
 |
Crystal Structure Of Tailspike Depolymerase (Apk14_Gp49) From Acinetobacter Phage Vb_Abap_Apk14
Organism: Acinetobacter phage vb_abap_apk14
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: PEG, GOL, CL |
 |
Crystal Structure Of A Tailspike Depolymerase (Apk16_Gp47) From Acinetobacter Phage Apk16
Organism: Acinetobacter phage apk16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-31 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Crystal Structure Of Tailspike Depolymerase (Apk09_Gp48) From Acinetobacter Phage Apk09
Organism: Acinetobacter phage apk09
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-05-31 Classification: VIRAL PROTEIN Ligands: PEG |
 |
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With The C-Terminal Part Of The T4 Gp5 Beta-Helical Domain
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: viral protein, hydrolase Ligands: MG, PLM, STE, ELA, ZN |
 |
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic The Phikz Central Spike Gp164
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, EDO, PLM, ZN |
 |
Photorhabdus Virulence Cassette (Pvc) Paar Repeat Protein Pvc10 In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic Pvc8, The Central Spike Protein Of Pvc
Organism: Enterobacteria phage t4, Photorhabdus luminescens subsp. laumondii (strain dsm 15139 / cip 105565 / tt01)
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, PLM |
 |
Chimera Of Bacteriophage Obp Gp146 Central Spike Protein And A T4 Gp5 Beta-Helix Fragment
Organism: Enterobacteria phage t4, Pseudomonas phage obp
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: FE2, STE, ELA, PLM, MG |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: UNKNOWN FUNCTION |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Apo Crystal Structure Of The Colanidase Tailspike Protein Gp150 Of Phage Phi92
Organism: Enterobacteria phage phi92
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: SR, CL, GOL, EDO |
 |
Crystal Structure Of The Colanidase Tailspike Protein Gp150 Of Phage Phi92 Complexed With One Repeating Unit Of Colanic Acid
Organism: Enterobacteria phage phi92
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: SO4, MG, EDO, PG4 |
 |
Organism: Acinetobacter phage vb_apip_p1
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: CL, NA |

