Search Count: 16
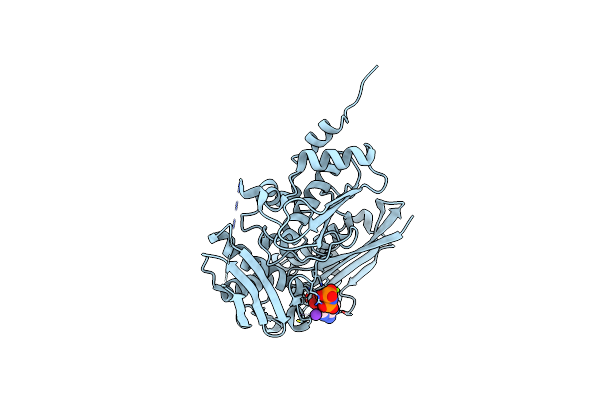 |
Mutant S-Adenosylmethionine Synthetase From E.Coli Complexed With Amppnp And Methionine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-13 Classification: TRANSFERASE Ligands: ANP, MG, K |
 |
S-Adenosylmethionine Synthetase From Lactobacillus Plantarum Complexed With Amppnp, Methionine And Sam
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2022-07-13 Classification: TRANSFERASE Ligands: PHE, PPK, MG, K, ANP, SAM |
 |
Organism: Escherichia coli (strain k12), Escherichia virus t3
Method: ELECTRON MICROSCOPY Release Date: 2021-07-21 Classification: HYDROLASE |
 |
Inter-Dimeric Interface Controls Function And Stability Of S-Methionine Adenosyltransferase From U. Urealiticum
Organism: Ureaplasma urealyticum serovar 7 str. atcc 27819
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-09-25 Classification: TRANSFERASE |
 |
Inter-Dimeric Interface Controls Function And Stability Of S-Methionine Adenosyltransferase From U. Urealiticum
Organism: Ureaplasma urealyticum serovar 7 str. atcc 27819
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-09-25 Classification: TRANSFERASE |
 |
Inter-Dimeric Interface Controls Function And Stability Of S-Methionine Adenosyltransferase From U. Urealiticum
Organism: Ureaplasma urealyticum serovar 7 str. atcc 27819
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: SAM, CL |
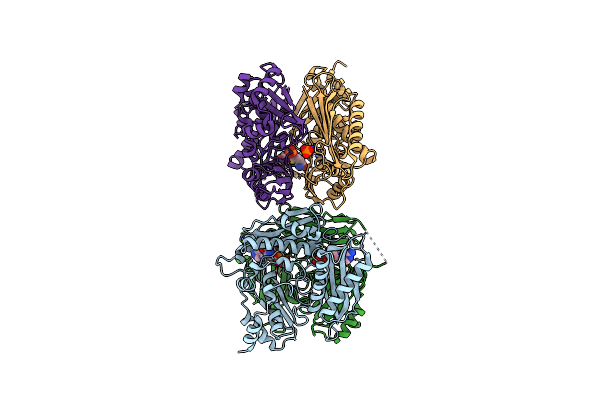 |
Inter-Dimeric Interface Controls Function And Stability Of S-Methionine Adenosyltransferase From U. Urealiticum
Organism: Ureaplasma urealyticum serovar 7 str. atcc 27819
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: ATP, SAM, PO4 |
 |
Inter-Dimeric Interface Controls Function And Stability Of S-Methionine Adenosyltransferase From U. Urealiticum
Organism: Ureaplasma urealyticum serovar 7 str. atcc 27819
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: K, MG, SAM, PPK |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: SO4, MN |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: SO4, MN |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1), Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: MN, SO4, MES |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1), Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: MN, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: BR, SO4 |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: CL, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: BR, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: CL, SO4 |

