Search Count: 28
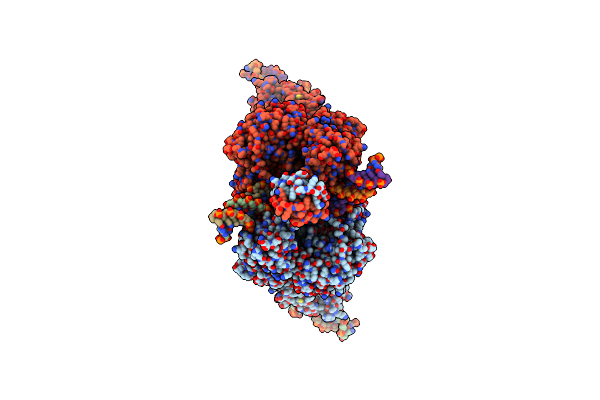 |
Cryo-Em Structure Of Tn4430 Tnpa Transposase From Tn3 Family In Complex With 100 Bp Long Transposon End Dna
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: RECOMBINATION |
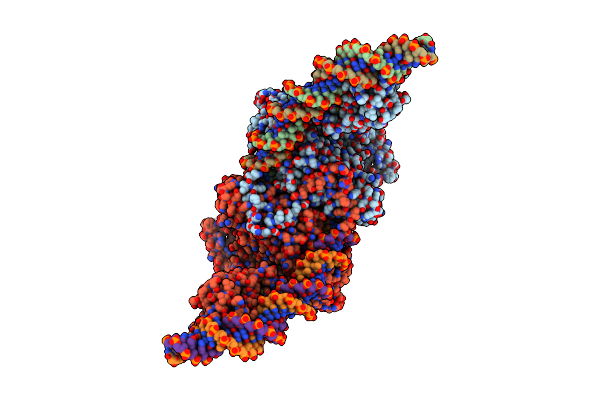 |
Cryo-Em Structure Of Tn4430 Tnpa Transposase From Tn3 Family In Complex With 48 Bp Long Transposon End Dna
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: RECOMBINATION |
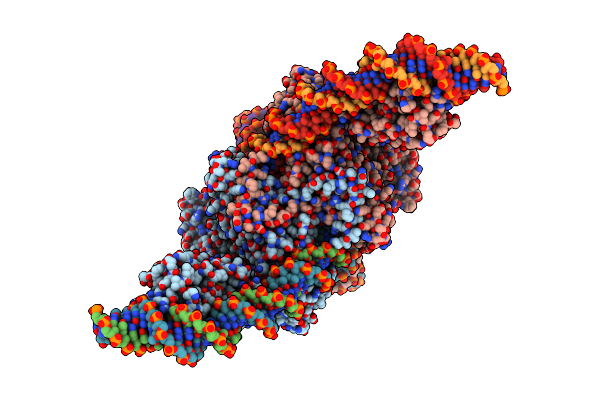 |
Cryo-Em Structure Of Tn4430 Tnpa Transposase From Tn3 Family In Complex With Strand-Transfer Like Dna Product
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: RECOMBINATION |
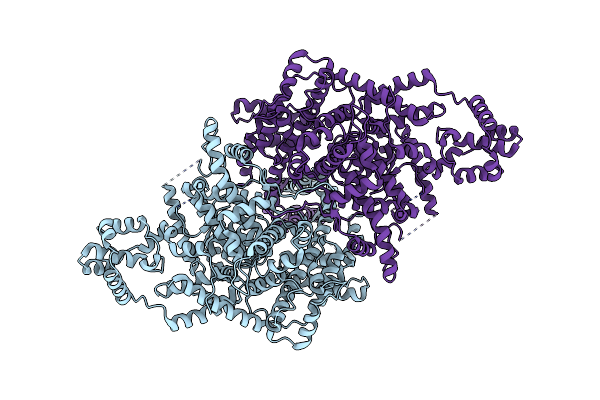 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: RECOMBINATION |
 |
Structural Basis Of Salm3 Dimerization And Adhesion Complex Formation With The Presynaptic Receptor Protein Tyrosine Phosphatases
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-07-22 Classification: CELL ADHESION Ligands: NAG |
 |
Crystal Structure Of An Asymmetric Dimer Of The N-Terminal Domain Of Euprosthenops Australis Major Ampullate Spidroin 1 (Dragline Silk)
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-17 Classification: PROTEIN FIBRIL Ligands: SO4 |
 |
Crystal Structure Of Mouse Salm5 Adhesion Protein Extracellular Lrr-Ig Domain Fragment
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-06-27 Classification: CELL ADHESION |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-26 Classification: LYASE Ligands: MG, NA, PO4 |
 |
Organism: Escherichia coli dh5[alpha]
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-01 Classification: HYDROLASE Ligands: GDP, MG |
 |
Organism: Escherichia coli o157:h7 str. ss52
Method: X-RAY DIFFRACTION Resolution:3.82 Å Release Date: 2016-08-24 Classification: TOXIN |
 |
Organism: Escherichia coli o157
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-06-01 Classification: TOXIN Ligands: GOL |
 |
Organism: Escherichia coli o157
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-06-01 Classification: TOXIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-03 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The N-Terminal Domain Of The Legionella Pneumophila Protein Sidc At 2.4A Resolution
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-02-26 Classification: UNKNOWN FUNCTION Ligands: MG, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-02-05 Classification: CHAPERONE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-02-05 Classification: CHAPERONE Ligands: GOL |
 |
Crystal Structure Of Trimeric Frataxin From The Yeast Saccharomyces Cerevisiae, Complexed With Ferrous
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-01-30 Classification: TRANSPORT PROTEIN Ligands: FE2 |
 |
Crystal Structure Of The Human Colony-Stimulating Factor 1 (Hcsf-1) Cytokine In Complex With The Viral Receptor Barf1
Organism: Human herpesvirus 4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2012-08-22 Classification: CYTOKINE RECEPTOR/CYTOKINE |
 |
Crystal Structure Of The Human Colony-Stimulating Factor 1 (Hcsf-1) Cytokine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-08-22 Classification: CYTOKINE |
 |
Crystal Structure Of The Mouse Colony-Stimulating Factor 1 (Mcsf-1) Cytokine
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-08-22 Classification: CYTOKINE Ligands: CA |

