Search Count: 20
 |
Organism: Echis carinatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: BLOOD CLOTTING Ligands: SO4 |
 |
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2015-11-04 Classification: TOXIN |
 |
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2015-08-26 Classification: TOXIN |
 |
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2015-08-26 Classification: TOXIN |
 |
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-08-13 Classification: TOXIN Ligands: SO4 |
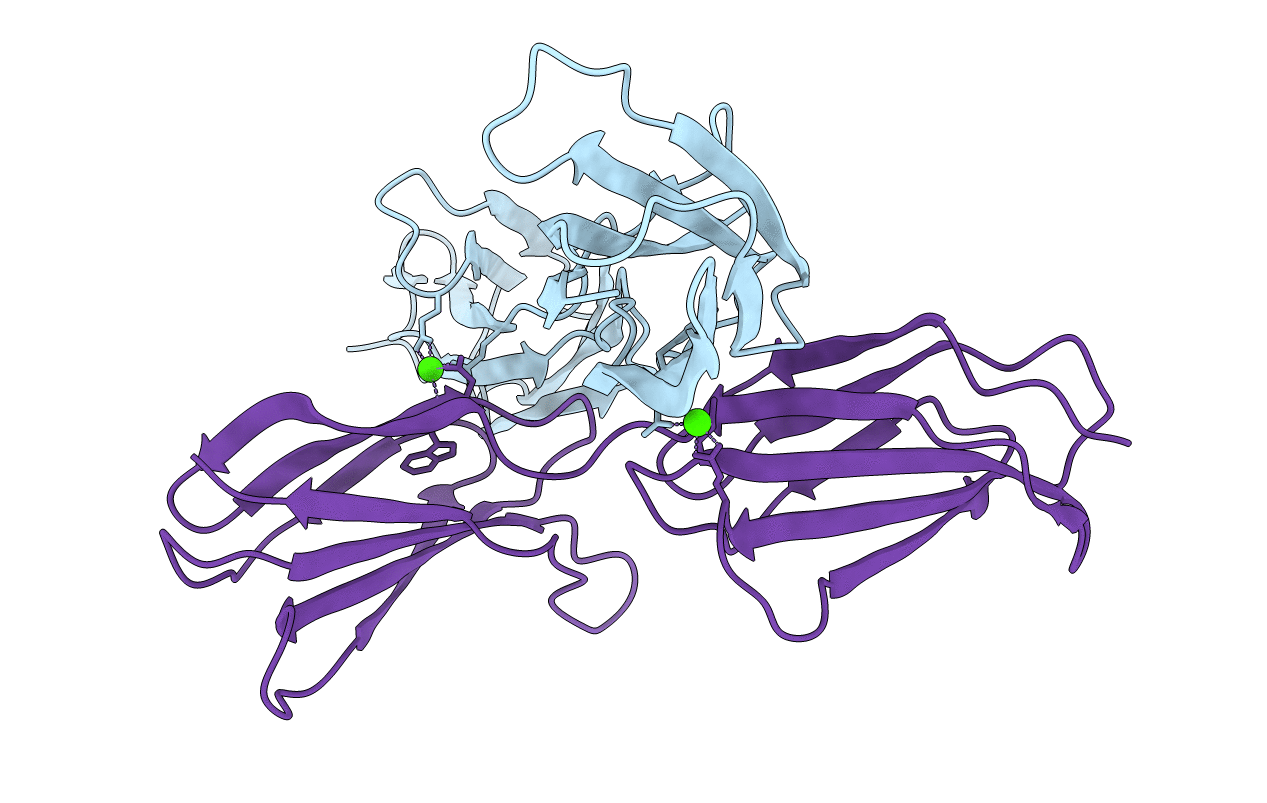 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2014-08-06 Classification: CELL ADHESION Ligands: CA |
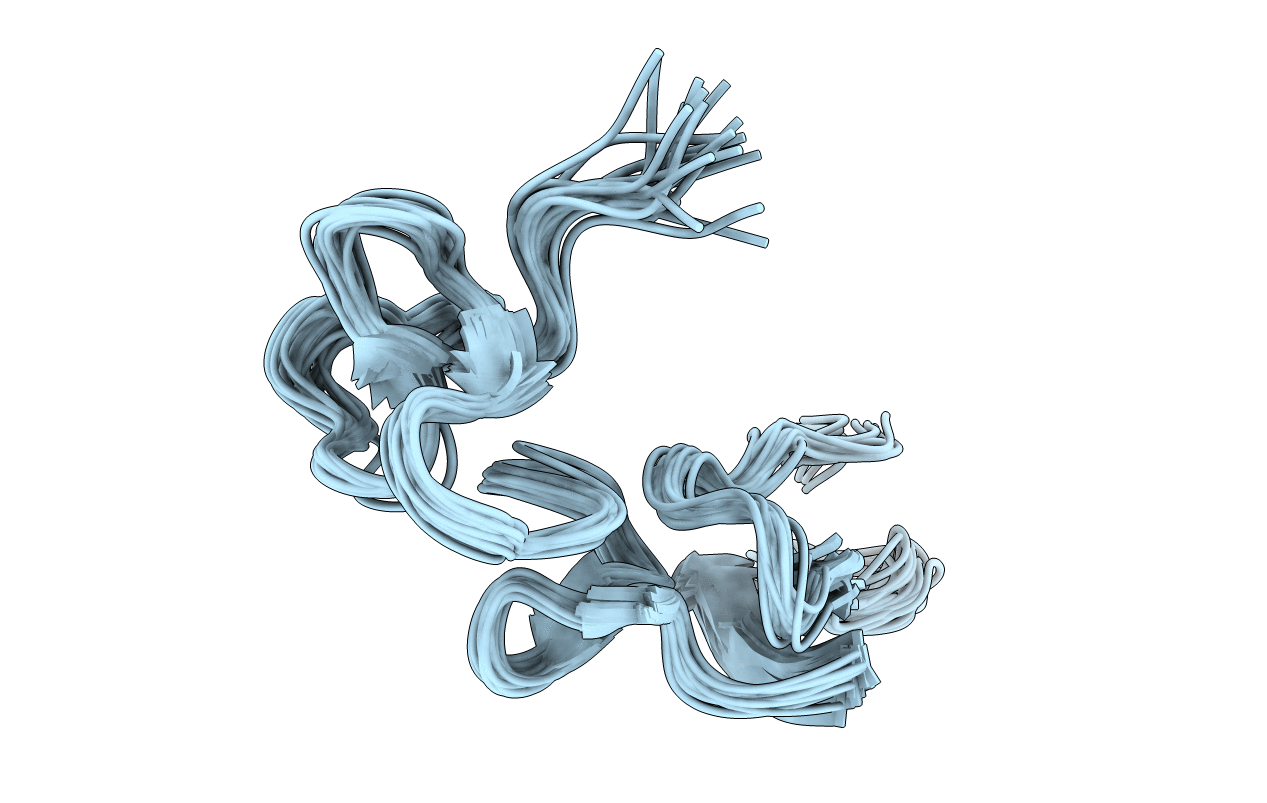 |
The C-Terminal Region Of Disintegrin Modulate Its 3D Conformation And Cooperate With Rgd Loop In Regulating Recognitions Of Integrins
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2013-05-22 Classification: HYDROLASE |
 |
The C-Terminal Region Of Disintegrin Modulate Its 3D Conformation And Cooperate With Rgd Loop In Regulating Integrins Recognitions
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2013-05-22 Classification: HYDROLASE |
 |
The C-Terminal Region Of Disintegrin Modulate Its 3D Conformation And Cooperate With Rgd Loop In Regulating Integrin Alpha-Iib Beta-3 Recognition
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2013-05-22 Classification: HYDROLASE |
 |
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2012-11-21 Classification: TOXIN |
 |
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2012-10-03 Classification: HYDROLASE |
 |
Expression In Pichia Pastoris And Backbone Dynamics Of Dendroaspin, A Three Finger Toxin
Organism: Dendroaspis jamesoni kaimosae
Method: SOLUTION NMR Release Date: 2012-03-07 Classification: TOXIN |
 |
Solution Structure And Backbone Dynamics Of The Dna-Binding Domain Of Foxp1: Insight Into Its Domain Swapping
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-04-21 Classification: DNA BINDING PROTEIN |
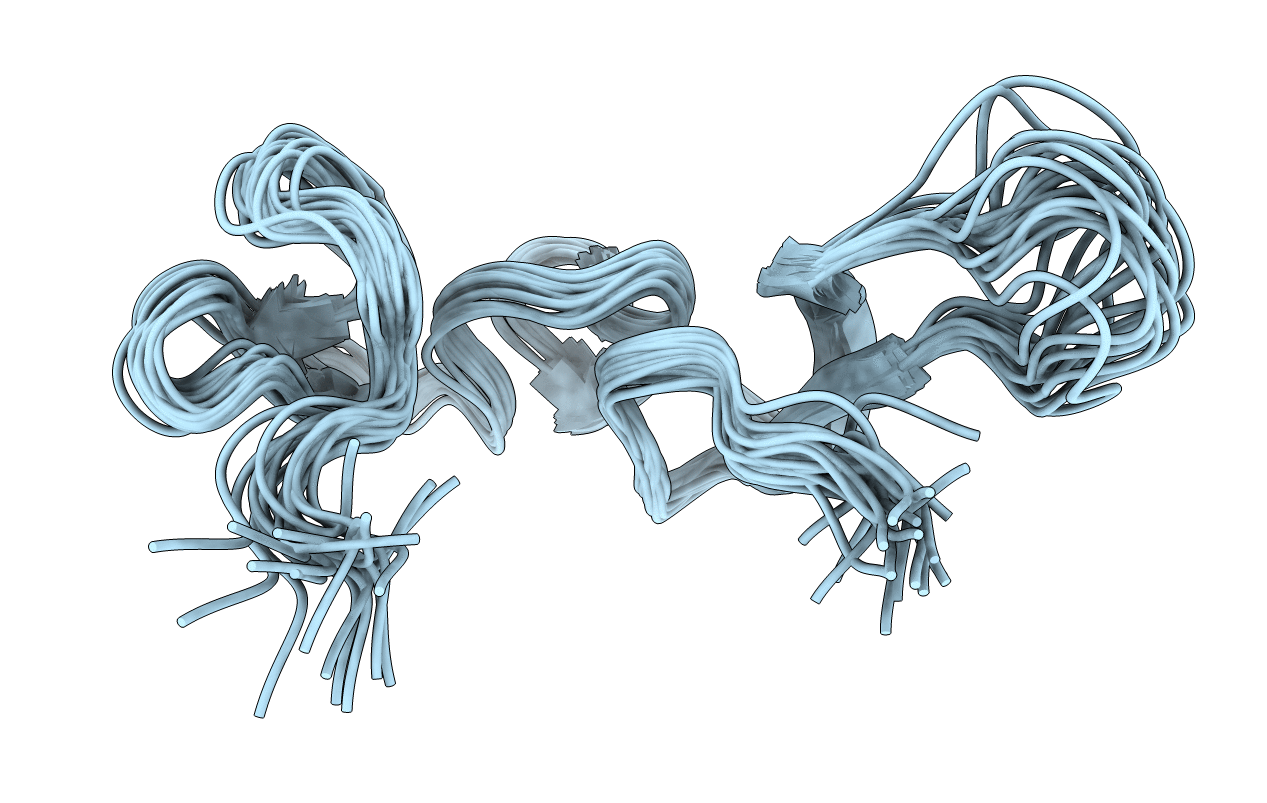 |
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2007-05-08 Classification: HYDROLASE |
 |
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2007-05-08 Classification: HYDROLASE |
 |
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2007-05-08 Classification: HYDROLASE |
 |
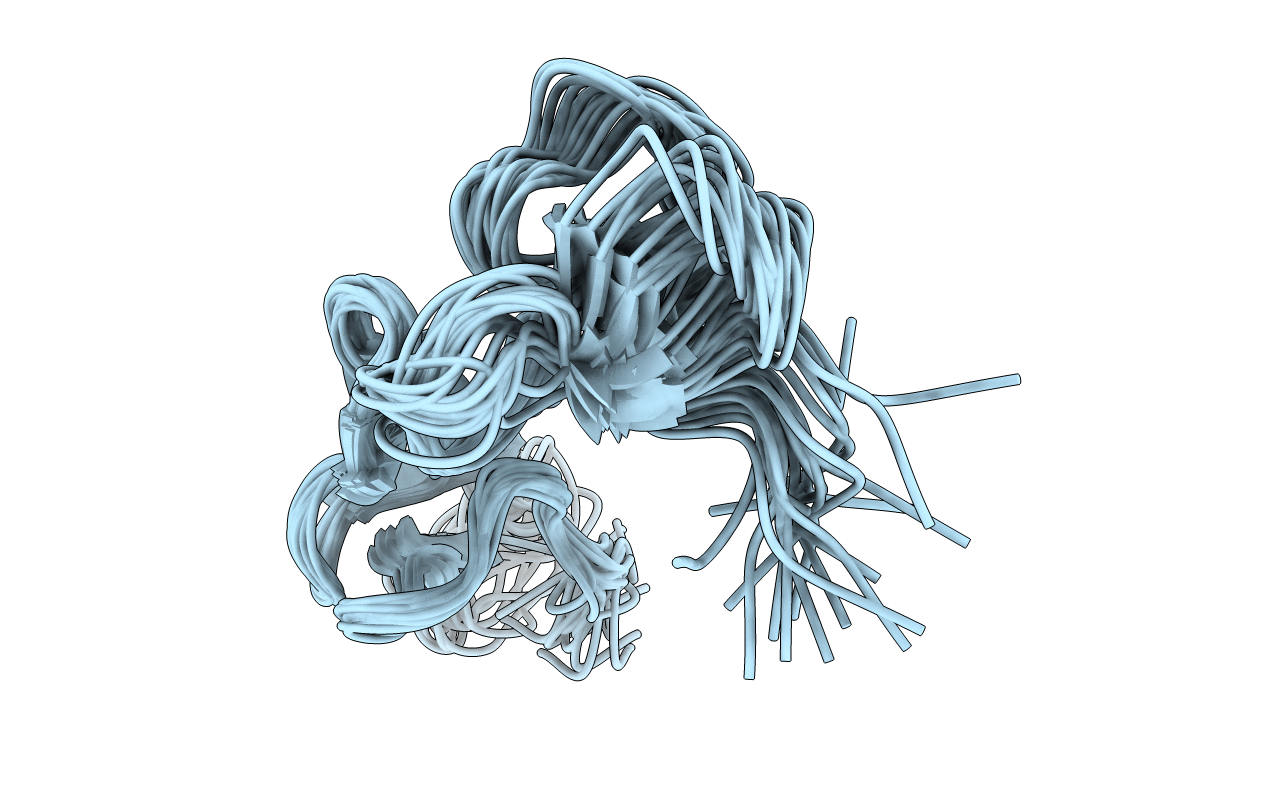
Structural Analysis Of Integrin Alpha Iib Beta 3- Disintegrin With The Akgdwn Motif
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2004-09-21 Classification: BLOOD CLOTTING, HYDROLASE |
 |
Structural Analysis Of Integrin Alpha Iib Beta 3- Disintegrin With The Akgdwn Motif
Organism: Calloselasma rhodostoma
Method: SOLUTION NMR Release Date: 2004-09-21 Classification: BLOOD CLOTTING, HYDROLASE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2004-05-18 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Solution Structure Of Gamma-Bungarotoxin:Implication For The Role Of The Residues Adjacent To Rgd In Integrin Binding
|

