Search Count: 132
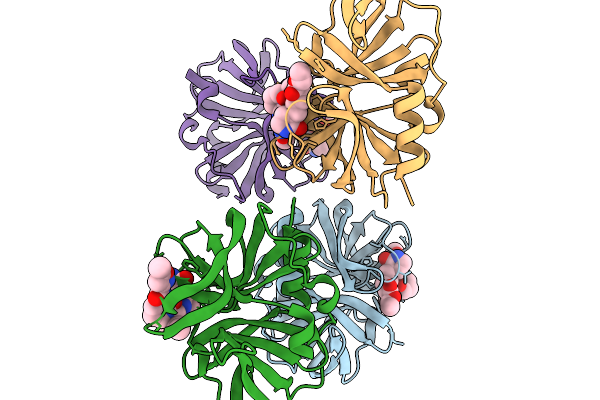 |
Organism: Rhinovirus b14
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: XNV |
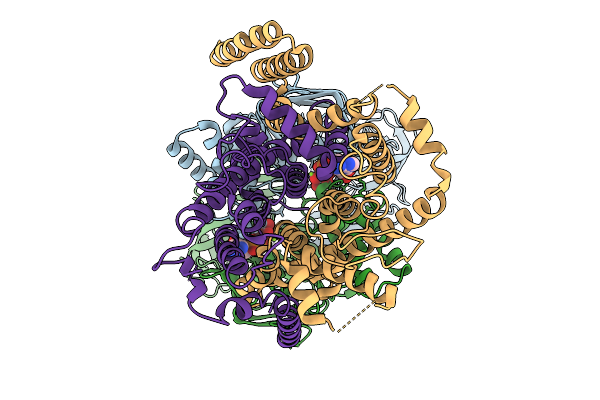 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN Ligands: MG, ADP, VO4 |
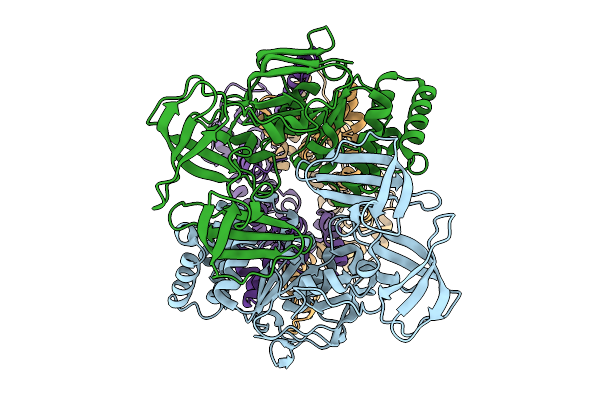 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN |
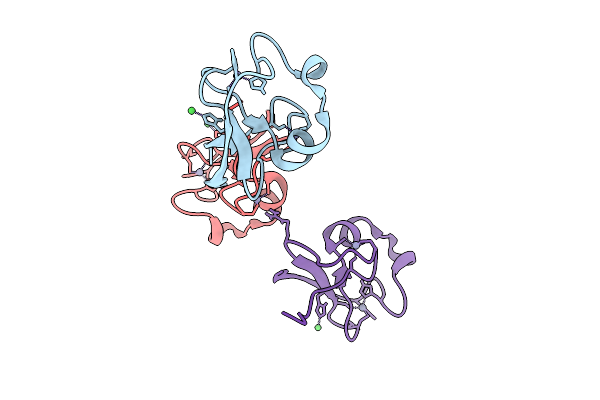 |
R-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Big
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: LIGASE Ligands: ZN, NI |
 |
Structure Of Canonical Myo7A-C Isoform (Adp-Bound) Expressed In Sensory Hair Cells (Head Domain + First Two Iq Domains), Bound To F-Actin
Organism: Mus musculus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, MG |
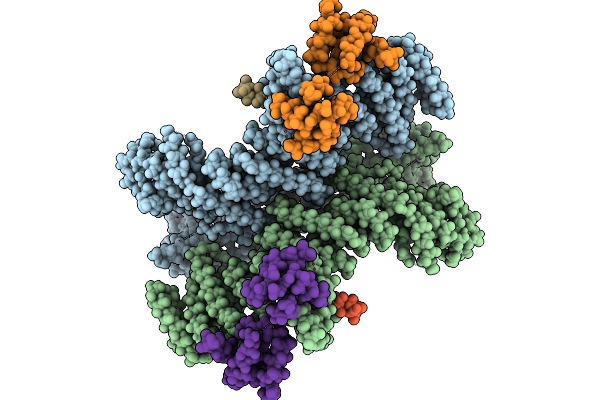 |
Cryo-Em Structure Of The Core Of The Arabidopsis Thaliana Ubr4/Di19/Calm1 Complex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
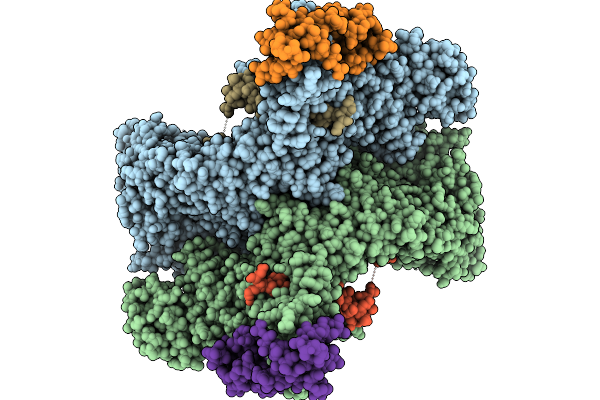 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
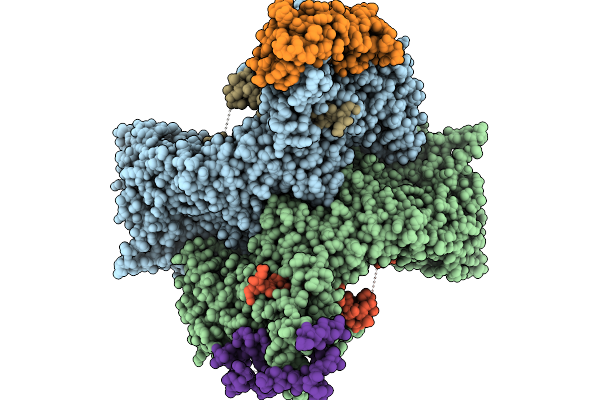 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Calm1 Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
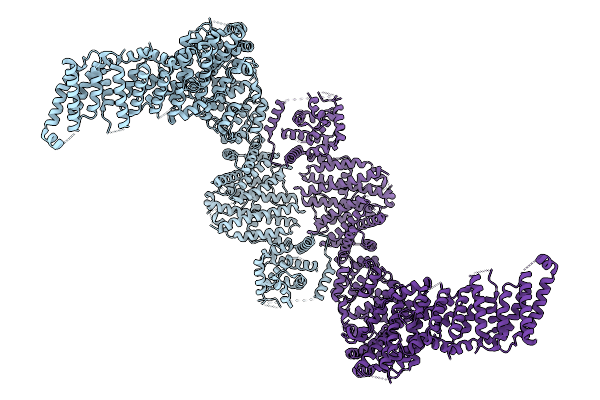 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (N-Term Focused Refinement)
|
 |
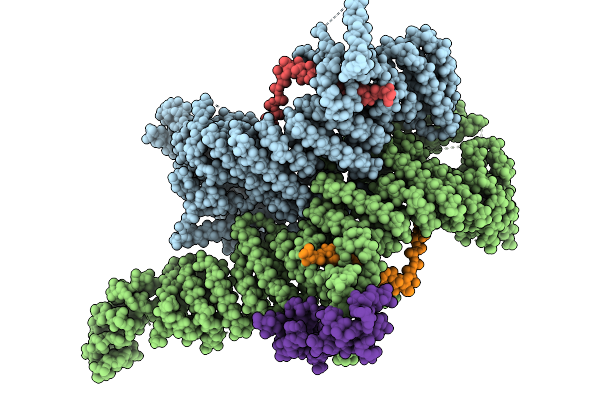
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Bp Focused Refinement)
|
 |
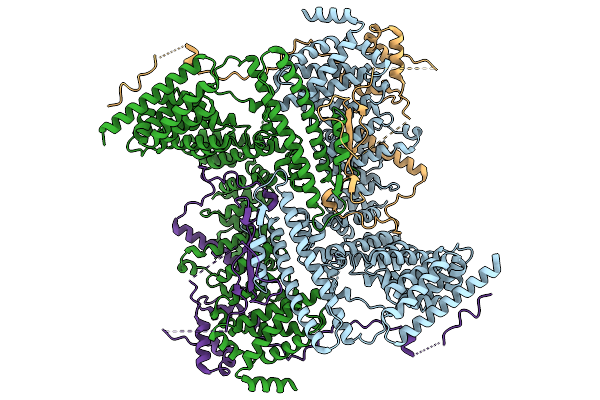
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Focused Refinement)
|
 |
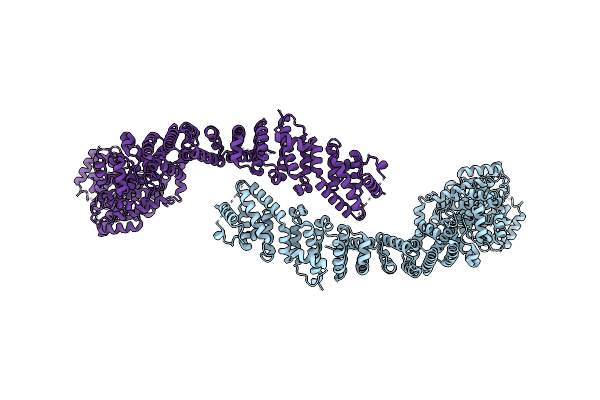
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
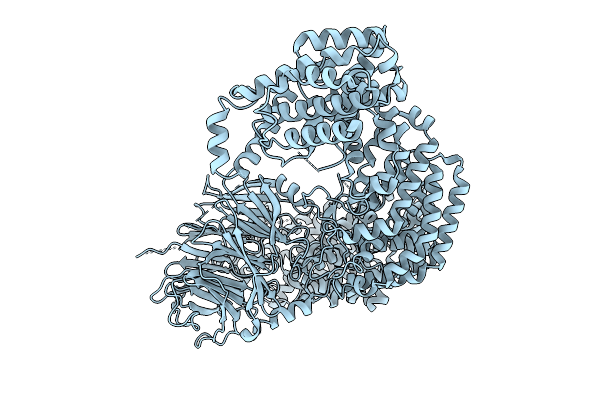
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (N-Term Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (Side Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: TRANSPORT PROTEIN |
 |
Structure Of A Mutated Photosystem Ii Complex Reveals Changes To The Hydrogen-Bonding Network That Affect Proton Egress During O-O Bond Formation
Organism: Synechocystis sp. pcc 6803
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, CLA, PHO, BCR, LMG, PL9, SQD, LMT, BCT, LHG, DGD, HEM, RRX, CA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-11-20 Classification: HYDROLASE/INHIBITOR Ligands: DMS, EDO, SO4, A1A21, A1AFD, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-11-20 Classification: HYDROLASE/INHIBITOR Ligands: A1AFE, DMS |

