Search Count: 27
 |
Fibrils Of The Super Helical Repeat Peptide, Shr-Ff, Grown At Elevated Temperature
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-06-27 Classification: DE NOVO PROTEIN |
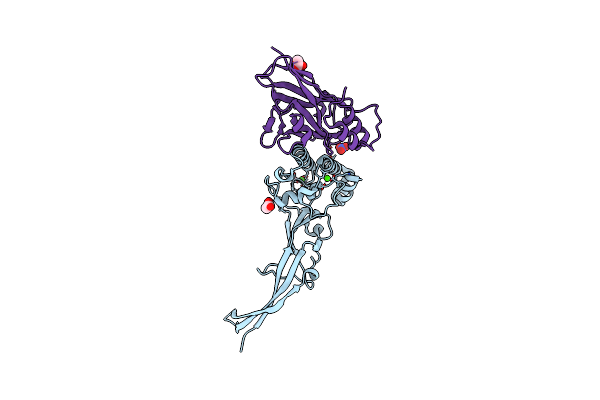 |
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2015-12-23 Classification: STRUCTURAL PROTEIN Ligands: CA, ACT, NO3 |
 |
Biomass Sensing Modules From Putative Rsgi-Like Proteins Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2013-09-11 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Biomass Sensoring Modules From Putative Rsgi-Like Proteins Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2013-09-11 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CA |
 |
Biomass Sensoring Module From Putative Rsgi2 Protein Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2013-09-11 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CA, ZN |
 |
High Resolution Structure For Family 3A Carbohydrate Binding Module From The Cipa Scaffolding Of Clostridium Thermocellum
Organism: Clostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2012-09-12 Classification: SUGAR BINDING PROTEIN Ligands: CA, SO4 |
 |
Carbohydrate-Binding Module Cbm3B From The Cellulosomal Cellobiohydrolase 9A From Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-04-25 Classification: HYDROLASE |
 |
Carbohydrate-Binding Module Cbm3B From The Cellulosomal Cellobiohydrolase 9A From Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2012-04-25 Classification: HYDROLASE Ligands: CA |
 |
Structure Of Cbm3B Of Major Scaffoldin Subunit Scaa From Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2012-01-11 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CA, EDO, NI, 1PE |
 |
Structure Of Cbm3B Of Major Scaffoldin Subunit Scaa From Acetivibrio Cellulolyticus Determined On The Nikel Absorption Edge
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-11 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, NI, 1PE |
 |
Structure Of Cbm3B Of Major Scaffoldin Subunit Scaa From Acetivibrio Cellulolyticus Determined From The Crystals Grown In The Presence Of Nickel
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2012-01-11 Classification: CRYSTALLINE CELLULOSE-BINDING PROTEIN Ligands: CA, EDO, NI, 1PE |
 |
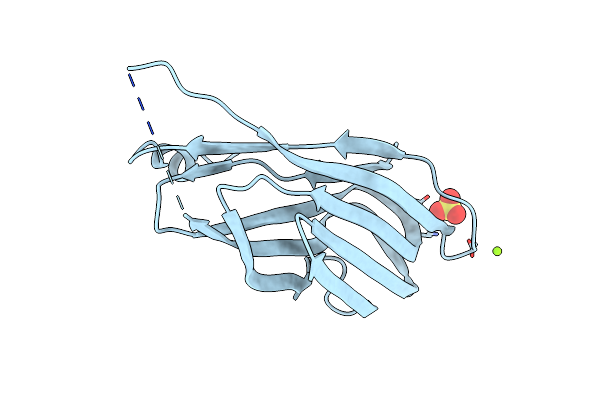
Structure Of A Scaffoldin Carbohydrate-Binding Module Family 3B From The Cellulosome Of Bacteroides Cellulosolvens: Structural Diversity And Implications For Carbohydrate Binding
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-04-06 Classification: SUGAR BINDING PROTEIN Ligands: NO3 |
 |
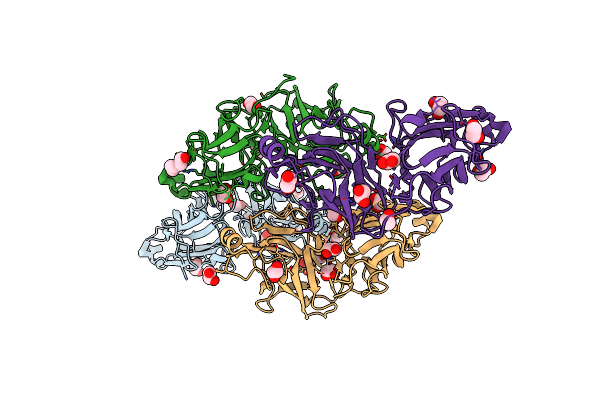
Non-Cellulosomal Cohesin From The Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2010-05-26 Classification: CELL ADHESION Ligands: CL, MG, SO4 |
 |
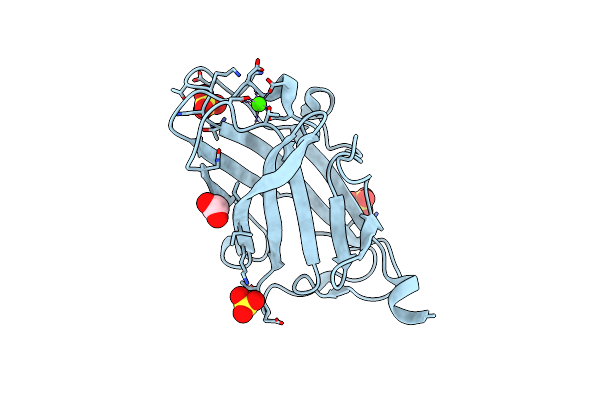
Structure Analysis Of The Type Ii Cohesin Dyad From The Adaptor Scaa Scaffoldin Of Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2010-05-05 Classification: STRUCTURAL PROTEIN, PROTEIN BINDING Ligands: EDO, PDO, HEZ |
 |
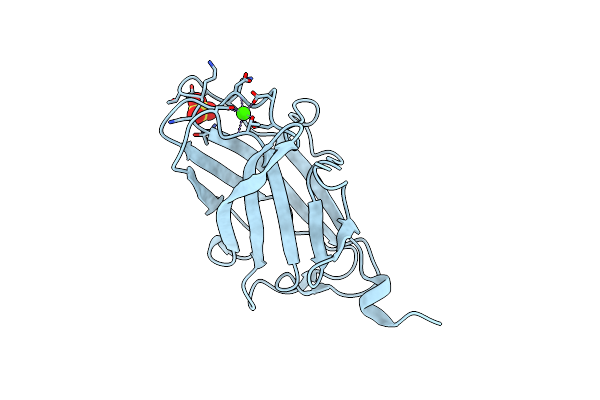
3B' Carbohydrate-Binding Module From The Cel9V Glycoside Hydrolase From Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2009-12-29 Classification: HYDROLASE Ligands: CA, SO4, FMT |
 |
3B' Carbohydrate-Binding Module From The Cel9V Glycoside Hydrolase From Clostridium Thermocellum, In-House Data
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-12-29 Classification: HYDROLASE Ligands: CA, SO4, CL |
 |
3B' Carbohydrate-Binding Module From The Cel9V Glycoside Hydrolase From Clostridium Thermocellum. Orthorhombic Structure
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-12-29 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Second Type Ii Cohesin Module From The Cellulosomal Adaptor Scaa Scaffoldin Of Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-06-23 Classification: STRUCTURAL PROTEIN Ligands: EDO, ACT, PDO, BU1 |
 |
Structure Of The Second Type Ii Cohesin Module From The Adaptor Scaa Scaffoldin Of Acetivibrio Cellulolyticus (Including Long C-Terminal Linker)
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2009-06-23 Classification: STRUCTURAL PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-01-20 Classification: TRANSCRIPTION Ligands: ZN |

