Search Count: 40
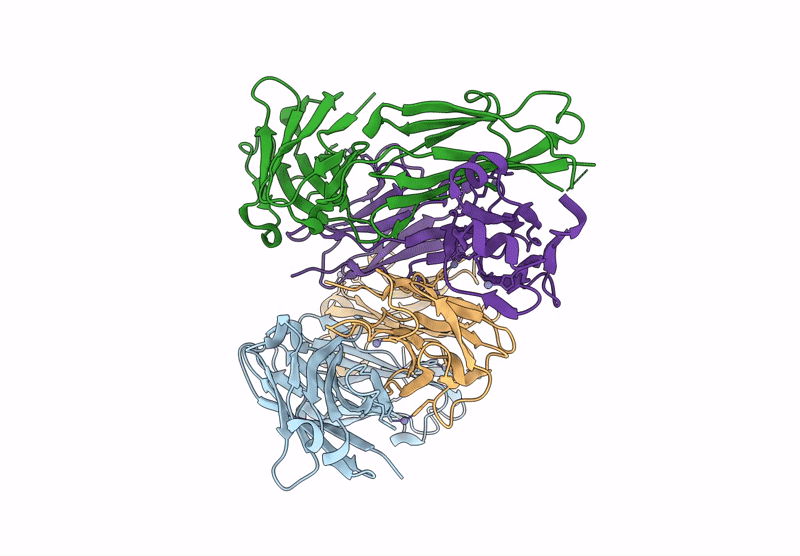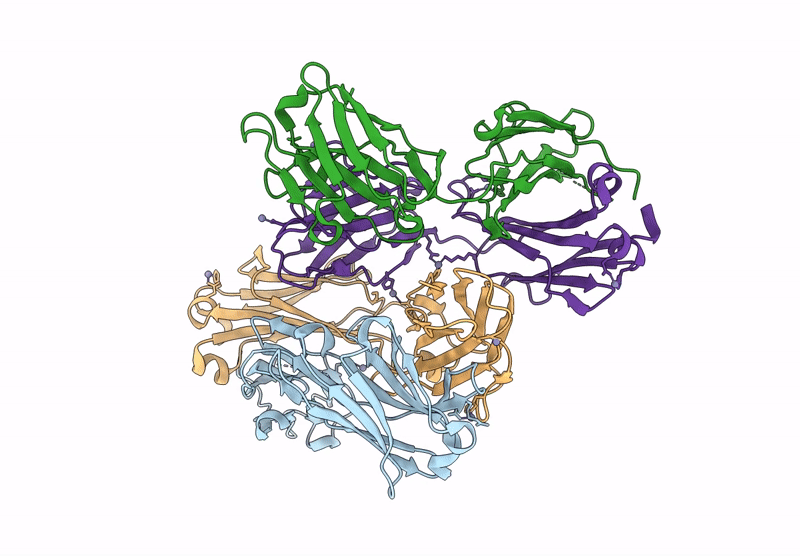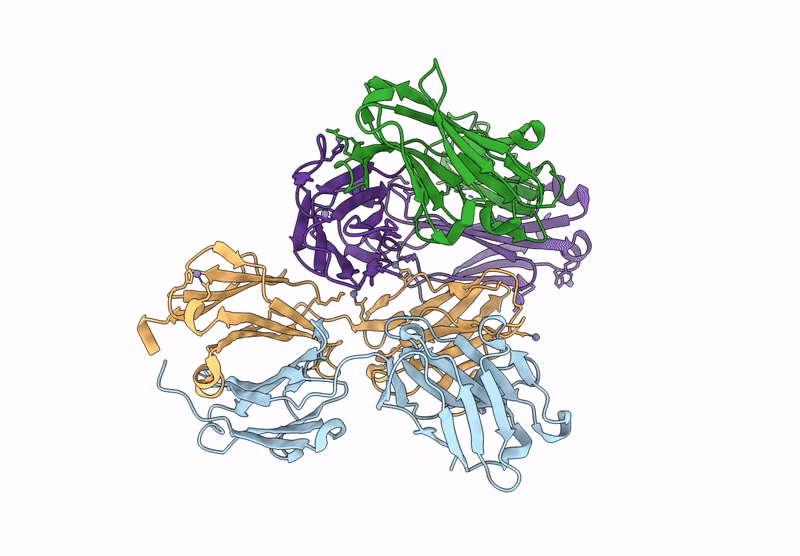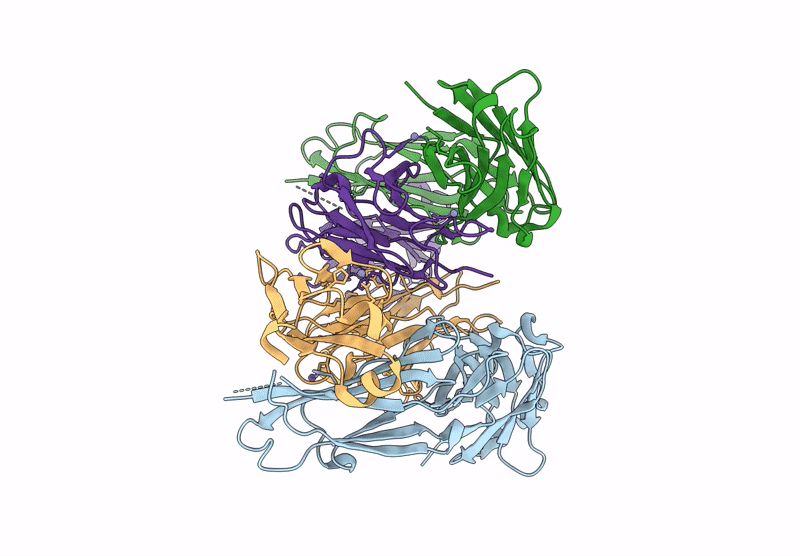 |
Crystal Structure Of Fab From Mouse Monoclonal Antibody 4A9 Against Aquifex Aeolicus Rsep Pdz Tandem
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-05 Classification: PROTEIN BINDING Ligands: ZN |
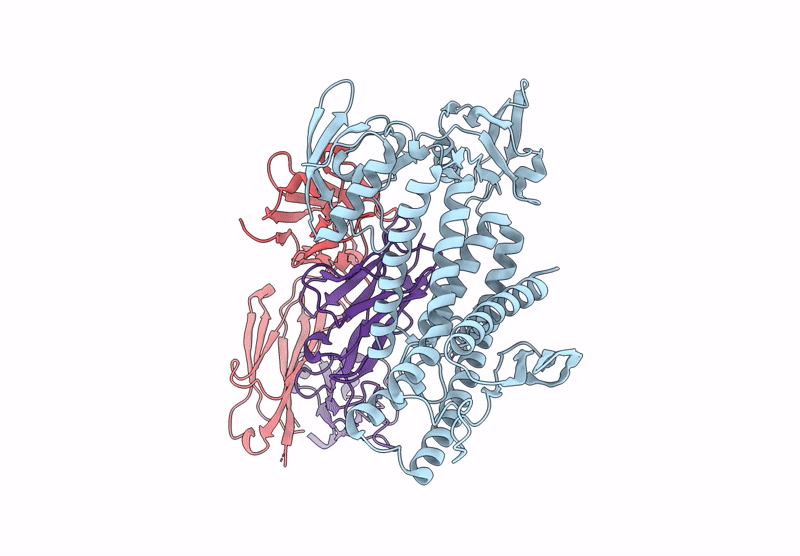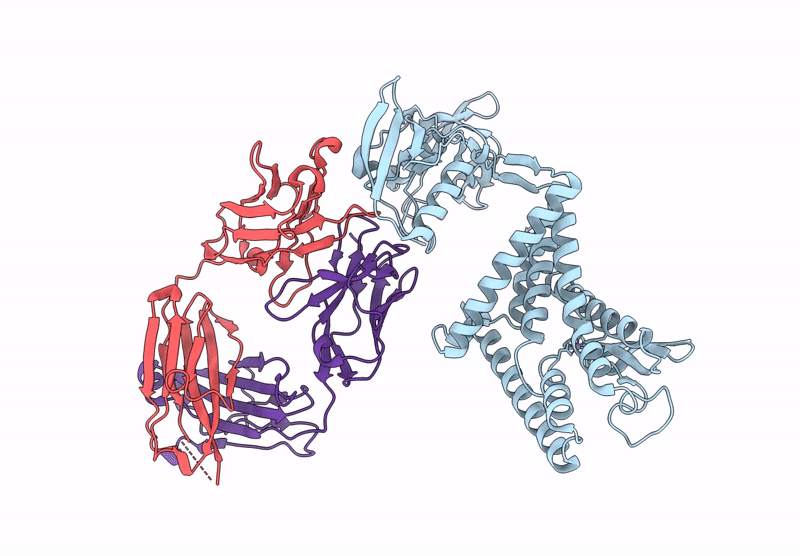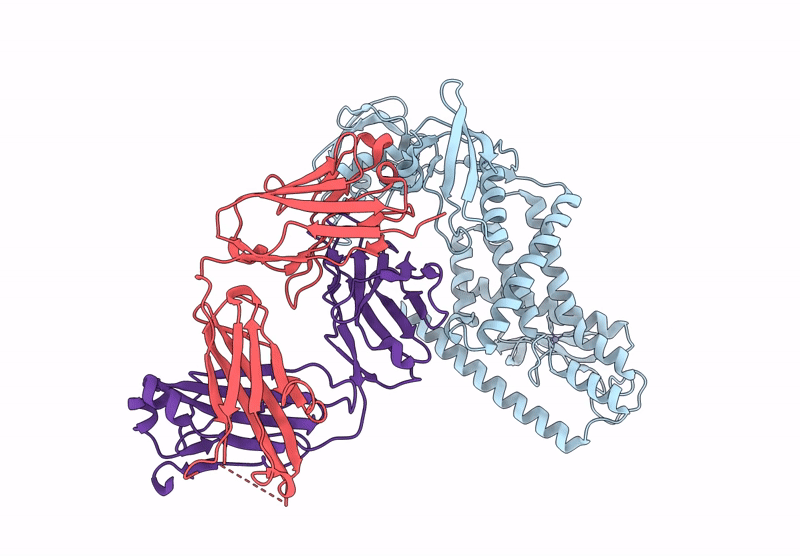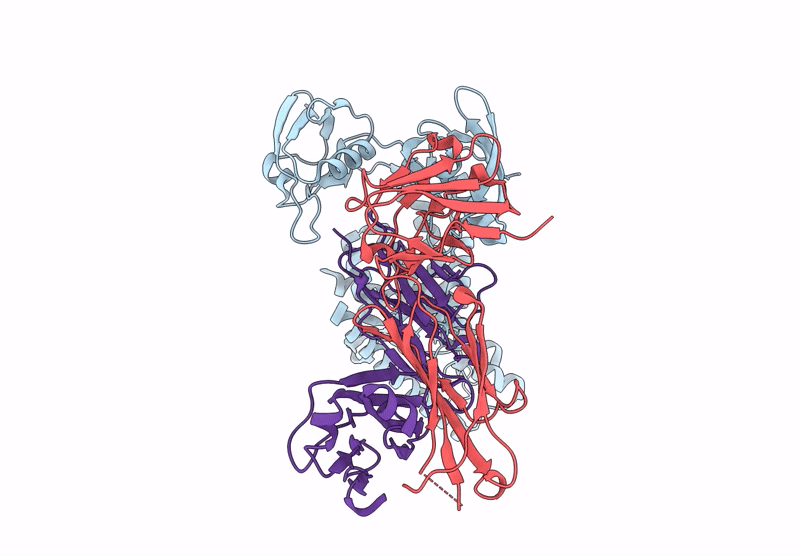 |
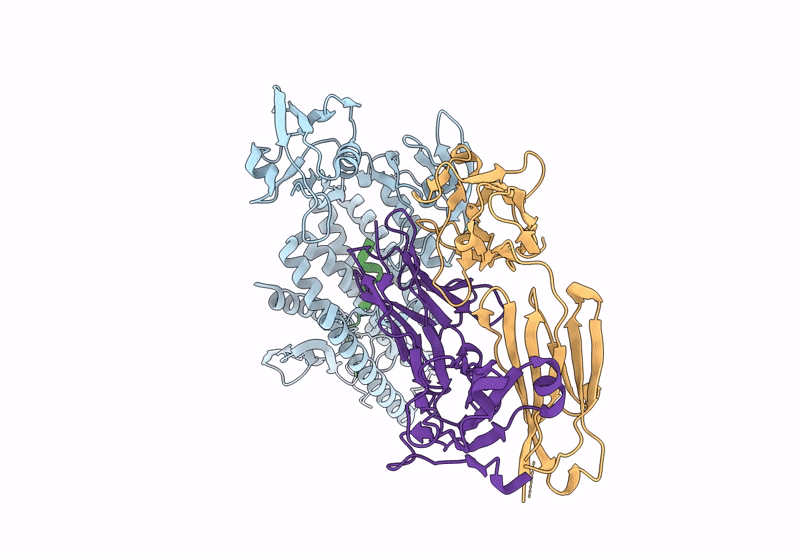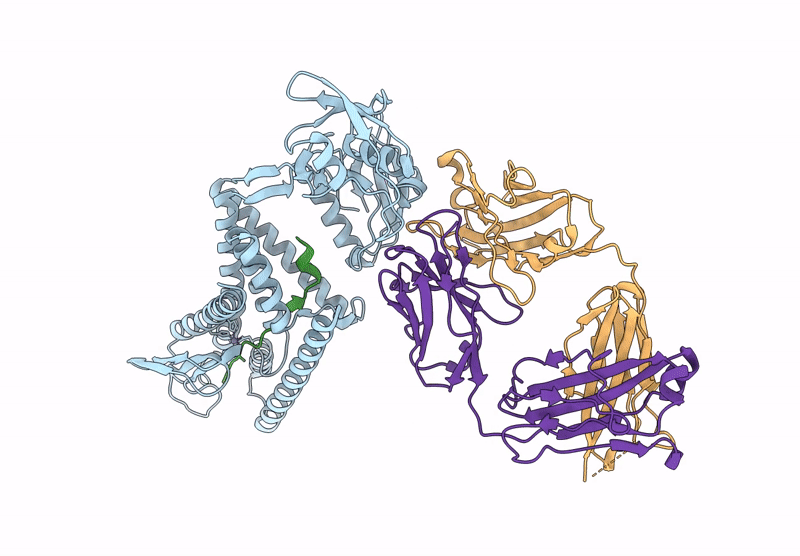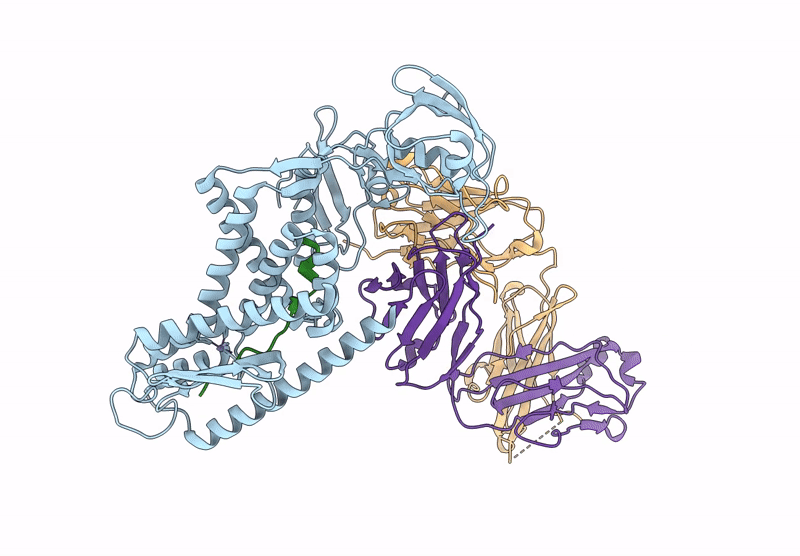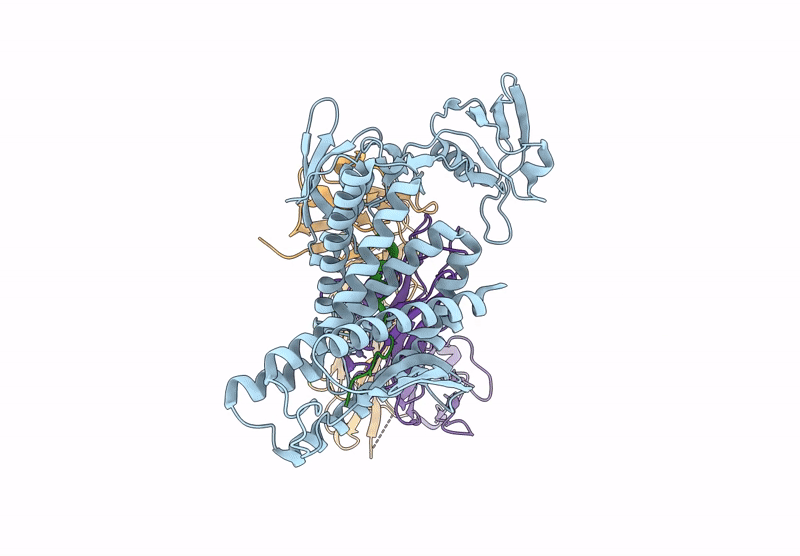
Organism: Aquifex aeolicus vf5, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Aquifex aeolicus vf5, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-09-21 Classification: HYDROLASE/INHIBITOR Ligands: T2L, DMS |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Inhibitor Tkb-248
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: T43, DMS, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: SIGNALING PROTEIN Ligands: A6F, OLA |
 |
Organism: Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: MEMBRANE PROTEIN Ligands: OLA, A6F |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-02-02 Classification: IMMUNE SYSTEM/DNA Ligands: EDO, SO4, IMD |
 |
Gamma-Glutamyltranspeptidase From Pseudomonas Nitroreducens Complexed With Gly-Gly
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: GLY, GOL |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-09-26 Classification: HYDROLASE Ligands: FLC, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-03-21 Classification: Transferase/Transferase Inhibitor Ligands: EJY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2018-03-21 Classification: Transferase/Transferase Inhibitor Ligands: EJP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2017-05-03 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Biotin At 1.3 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: BTN, SO4 |
 |
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Biotin Long Tail (Btntail) At 1.5 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOE, GOL, CD, P6G |
 |
Crystal Structure Of The Core Streptavidin Mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) Complexed With Iminobiotin Long Tail (Imntail) At 1.2 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOF, GOL |
 |
Crystal Structure Of The Core Streptavidin Mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) Complexed With Iminobiotin Long Tail (Imntail) At 1.7 A Resolution
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-18 Classification: BIOTIN BINDING PROTEIN Ligands: ZOF, P6G |

