Search Count: 48
 |
Organism: Nostoc sp.
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, LMT, LMG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-01-13 Classification: IMMUNE SYSTEM Ligands: ATP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2021-01-13 Classification: IMMUNE SYSTEM Ligands: SO4, ATP |
 |
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-08-14 Classification: TRANSPORT PROTEIN Ligands: SLB |
 |
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-08-14 Classification: TRANSPORT PROTEIN Ligands: SLB, CL, CS |
 |
Organism: Pichia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Hetero-Trimeric Core Of Lubac: Hoip Double-Uba Complexed With Hoil-1L Ubl And Sharpin Ubl
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-05-02 Classification: LIGASE Ligands: GOL |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of Old Yellow Enzyme From Candida Macedoniensis Aku4588 Complexed With P-Hydroxybenzaldehyde
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN, HBA |
 |
Crystal Structure Of Low-Specificity L-Threonine Aldolase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-12-17 Classification: LYASE Ligands: PLG |
 |
Crystal Structure Of Nadph-Dependent 3-Quinuclidinone Reductase From Rhodotorula Rubra
Organism: Rhodotorula mucilaginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-08-13 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG, GLY |
 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG |
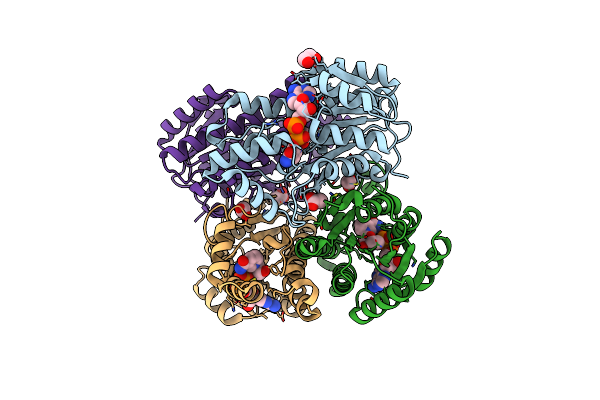 |
Crystal Structure Of 3-Quinuclidinone Reductase From Agrobacterium Tumefaciens
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2014-06-25 Classification: OXIDOREDUCTASE Ligands: NAD, ACY, EDO |
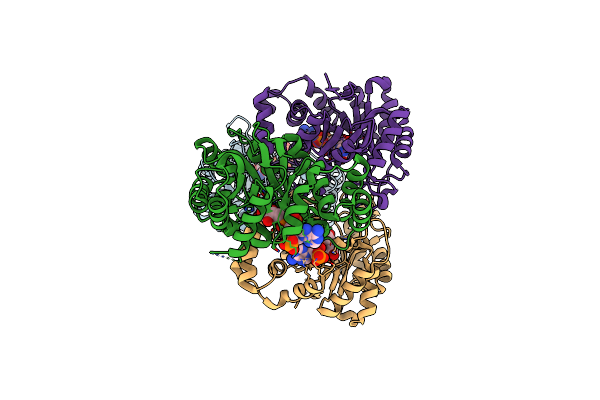 |
Crystal Structure Of Conjugated Polyketone Reductase C1 From Candida Parapsilosis Complexed With Nadph
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-08-21 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
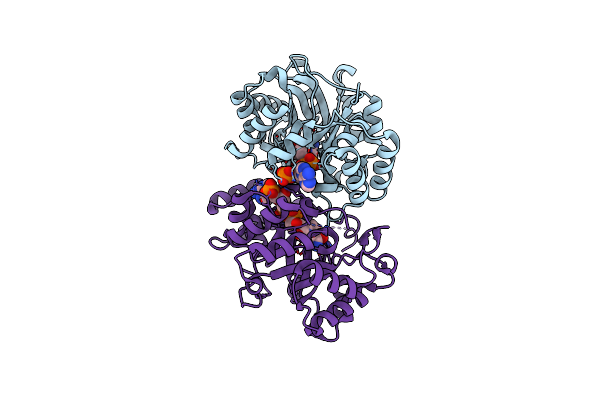
Crystal Structure Of Conjugated Polyketone Reductase C2 From Candida Parapsilosis
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE |
 |
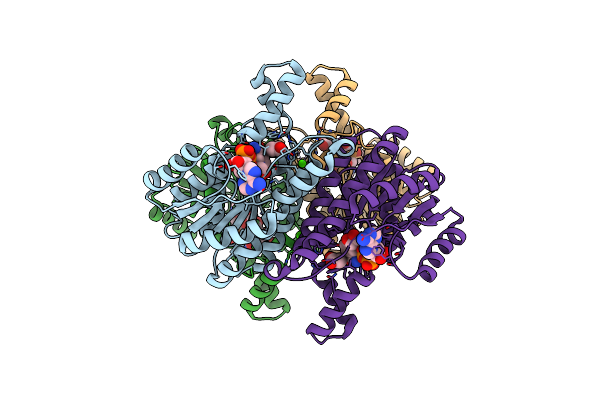
Crystal Structure Of Conjugated Polyketone Reductase C2 From Candida Parapsilosis Complexed With Nadph
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Alcaligenes Faecalis D-3-Hydroxybutyrate Dehydrogenase In Complex With Nad(+) And Acetate
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-02-29 Classification: OXIDOREDUCTASE Ligands: ACT, CL, NAD, CA |
 |
Crystal Structure Of D-3-Hydroxybutyrate Dehydrogenase, Prepared In The Presence Of The Substrate D-3-Hydroxybutyrate And Nad(+)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-02-08 Classification: OXIDOREDUCTASE Ligands: CA, CL, NAD, NAI, 3HR, AAE |
 |
The Unique Structure Of Wild Type Carbonic Anhydrase Alpha-Ca1 From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2011-11-16 Classification: LYASE Ligands: ZN, SO4, NAG |

