Search Count: 47
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of Sars-Cov-2 Xbb.1.5 Spike Glycoprotein In Complex With Ace2 (1-Up State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Structure Of Sars-Cov-2 Xbb.1.5 Spike Glycoprotein In Complex With Ace2 (2-Up State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2023-09-27 Classification: FLUORESCENT PROTEIN |
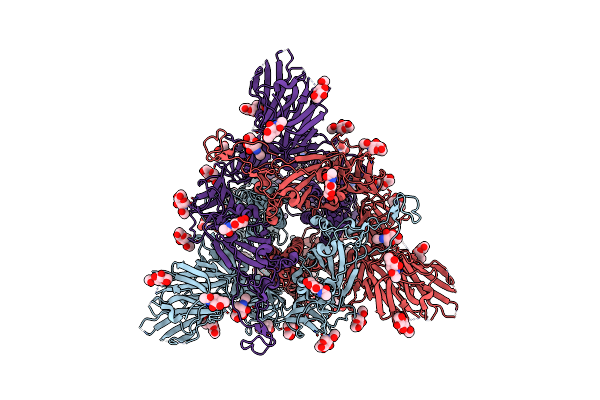 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: VIRAL PROTEIN Ligands: NAG |
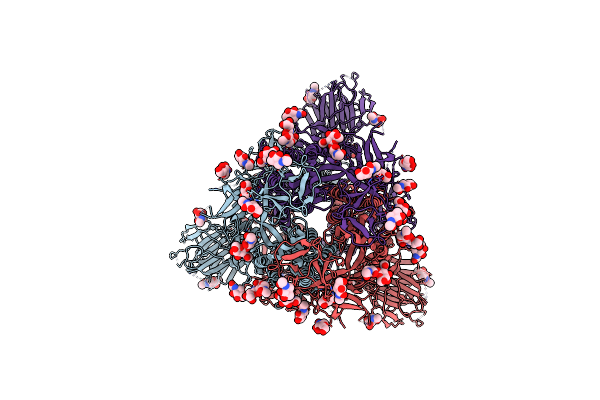 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: VIRAL PROTEIN Ligands: NAG |
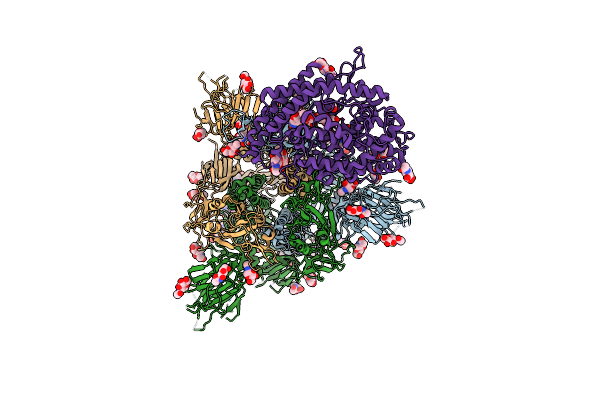 |
Structure Of Sars-Cov-2 Xbb.1 Spike Glycoprotein In Complex With Ace2 (1-Up State)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
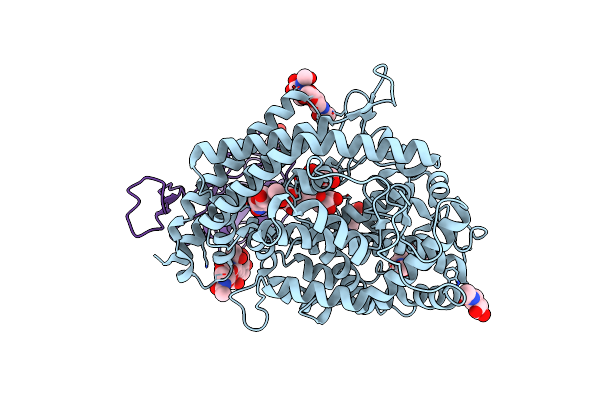 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: VIRAL PROTEIN/PROTEIN BINDING Ligands: NAG |
 |
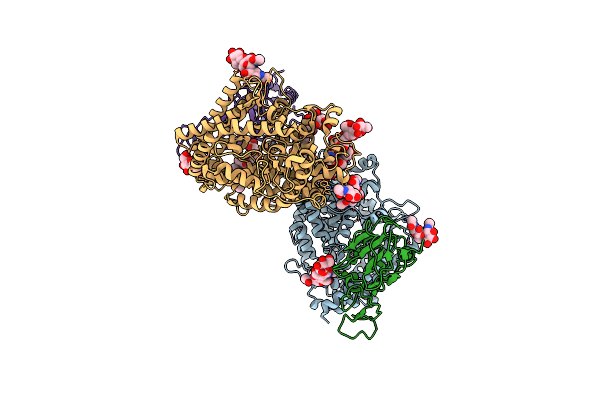
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
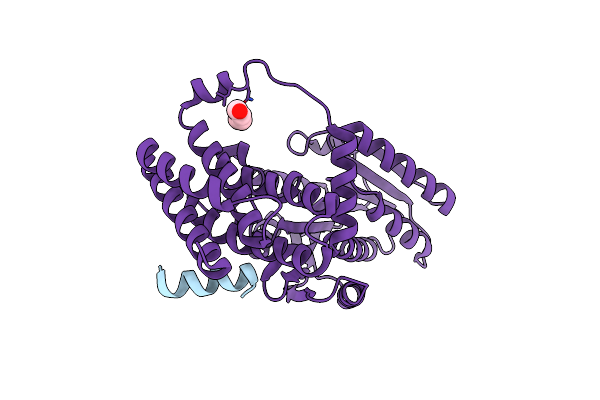
Crystal Structure Of The Receptor Binding Domain Of Sars-Cov-2 Omicron Ba.4/5 Variant Spike Protein In Complex With Its Receptor Ace2
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2022-09-28 Classification: VIRAL PROTEIN Ligands: ZN, NAG |
 |
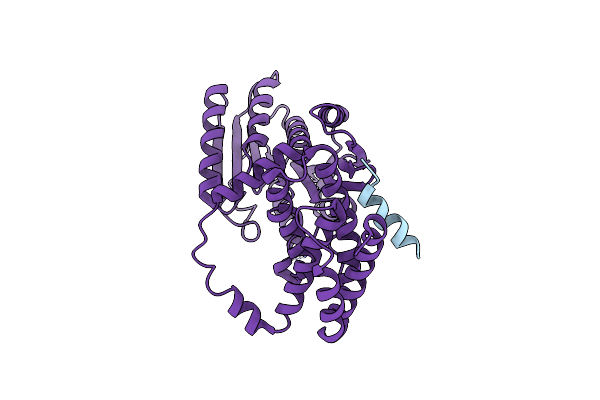
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-09-02 Classification: TRANSCRIPTION Ligands: PEG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-09-02 Classification: TRANSCRIPTION |
 |
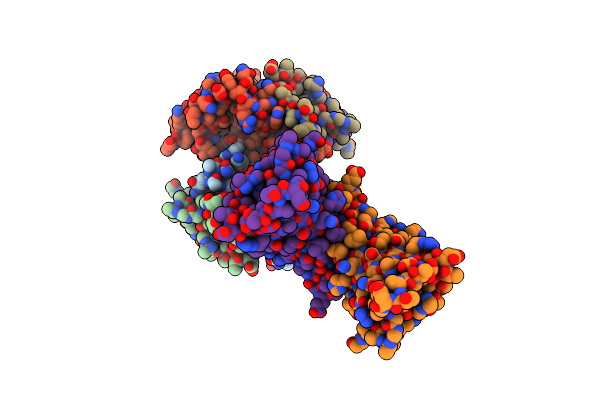
Organism: Aequorea victoria
Method: NEUTRON DIFFRACTION Resolution:1.44 Å Release Date: 2020-04-08 Classification: FLUORESCENT PROTEIN Ligands: D3O, DOD |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.77 Å Release Date: 2020-04-01 Classification: FLUORESCENT PROTEIN Ligands: DOD |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-18 Classification: PEPTIDE BINDING PROTEIN Ligands: GOL |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE |
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2018-12-26 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-12-26 Classification: DNA BINDING PROTEIN Ligands: SO4 |

