Search Count: 104
 |
Structure Of Beta-1,2-Glucanase From Endozoicomonas Elysicola (Eesgl1, Ligand-Free)
Organism: Endozoicomonas elysicola dsm 22380
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Structure Of Beta-1,2-Glucanase From Photobacterium Gaetbulicola (Pgsgl3, Ligand-Free)
Organism: Photobacterium gaetbulicola gung47
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CL |
 |
Structure Of Beta-1,2-Glucanase From Xanthomonas Campestris Pv. Campestris (Beta-1,2-Glucoheptasaccharide Complex)-E239Q Mutant
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
 |
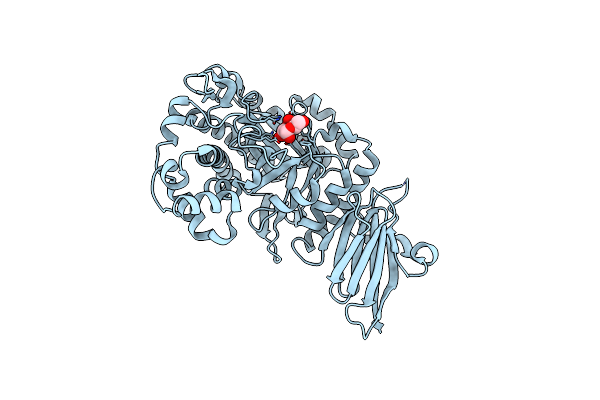
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL |
 |
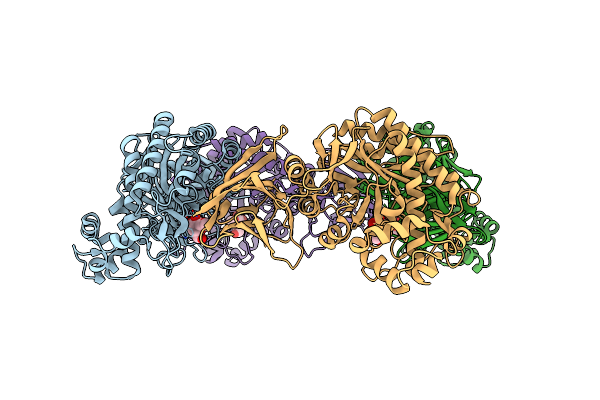
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GLA |
 |
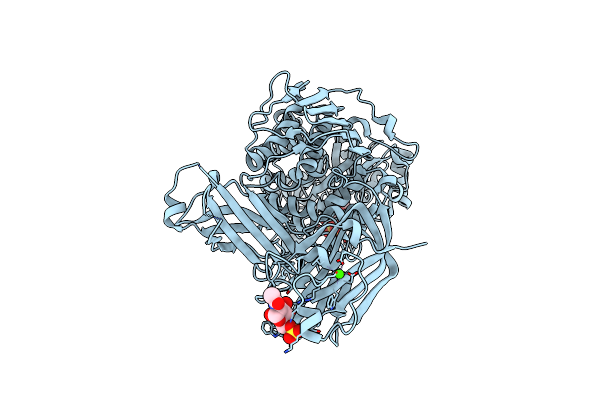
Beta-Galactosidase From Bacteroides Xylanisolvens (Complex With Methyl Beta-Galactopyranose)
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: MBG |
 |
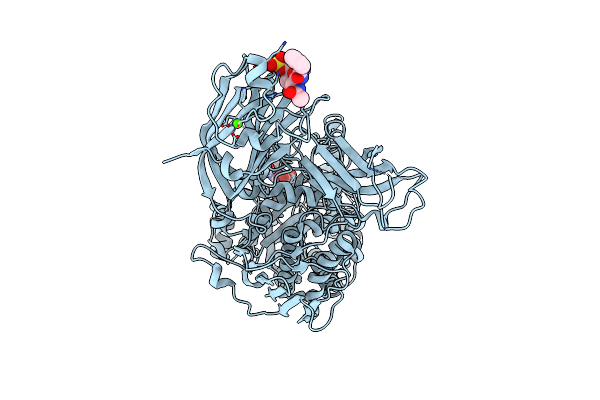
6-Sulfo-Beta-D-N-Acetylglucosaminidase From Bifidobacterium Bifidum In Complex With Glcnac-6S
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: NGS, NGY, CA |
 |
6-Sulfo-Beta-D-N-Acetylglucosaminidase From Bifidobacterium Bifidum In Complex With Pugnac-6S
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: 8R9, CA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-14 Classification: RNA Ligands: ZN, SO4, GOL, EDO, ACT |
 |
Pholiota Squarrosa Lectin (Phosl) In Complex With Fucose(Alpha1-6)[Glcnac(Beta1-4)]Glcnac
Organism: Pholiota squarrosa
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-10-12 Classification: SUGAR BINDING PROTEIN Ligands: FMT |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Sophotetraose Observed As Sophorose
Organism: Ignavibacterium album
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: CA |
 |
The Apo Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-03-09 Classification: TRANSFERASE |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Glucose
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: BGC, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Sophorose
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With 1-Deoxynojirimycin
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: NOJ, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With P-Nitrophenyl-Alpha-D-Glucopyranoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: PNG, CA |
 |
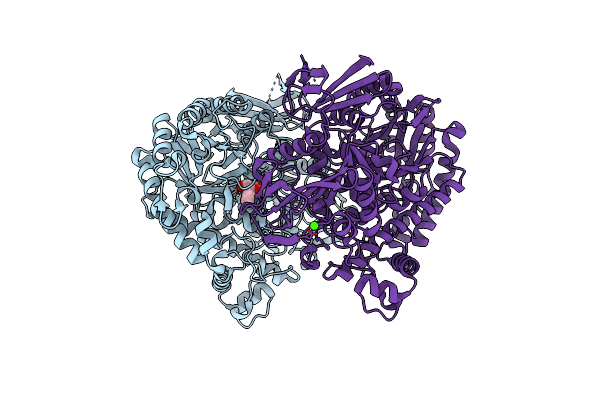
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Methyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: GYP, CA |
 |
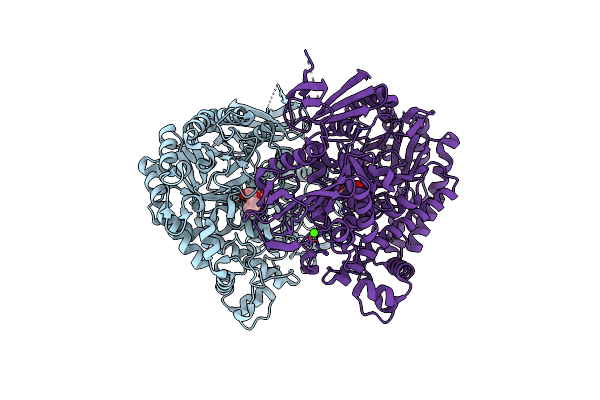
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Ethyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: BW3, CA |
 |
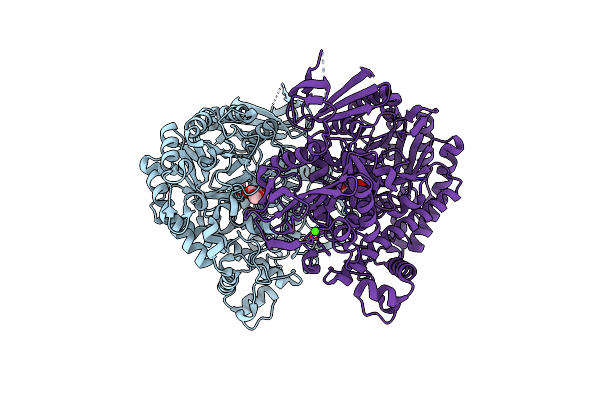
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Phenyl Alpha-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 7OL, CA |
 |
The Complex Structure Of Beta-1,2-Glucosyltransferase From Ignavibacterium Album With Methyl Beta-D-Glucoside
Organism: Ignavibacterium album (strain dsm 19864 / jcm 16511 / nbrc 101810 / mat9-16)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: MGL, CA |

