Search Count: 7
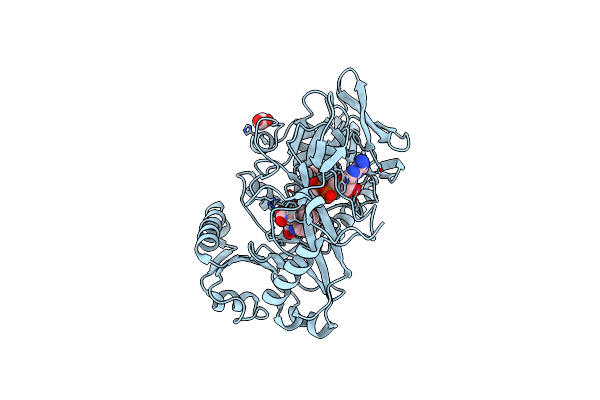 |
Organism: Phaeosphaeria nodorum (strain sn15 / atcc mya-4574 / fgsc 10173)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-06-28 Classification: OXIDOREDUCTASE Ligands: FAD, ACY |
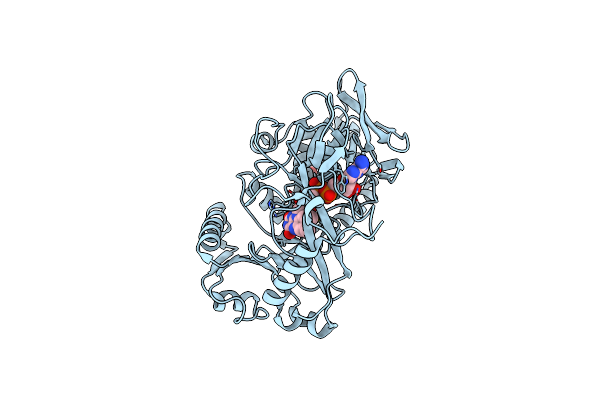 |
Crystal Structure Of Phaeospaeria Nodrum Fructosyl Peptide Oxidase Mutant Asn56Ala
Organism: Phaeosphaeria nodorum (strain sn15 / atcc mya-4574 / fgsc 10173)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-06-28 Classification: OXIDOREDUCTASE Ligands: FAD, ACY |
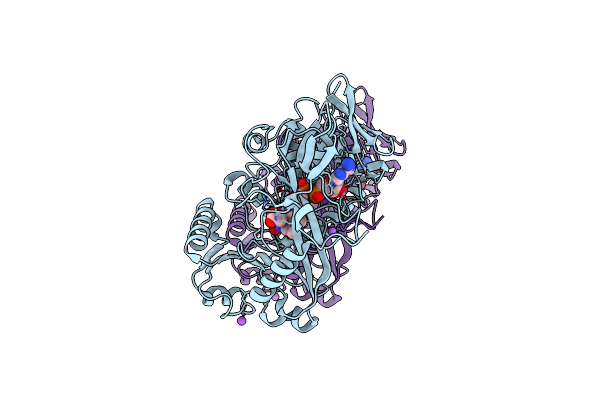 |
Crystal Structure Of Phaeospaeria Nodrum Fructosyl Peptide Oxidase Mutant Asn56Ala In Complexes With Sodium And Chloride Ions
Organism: Phaeosphaeria nodorum (strain sn15 / atcc mya-4574 / fgsc 10173)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-28 Classification: OXIDOREDUCTASE Ligands: FAD, ACY, CL, NA |
 |
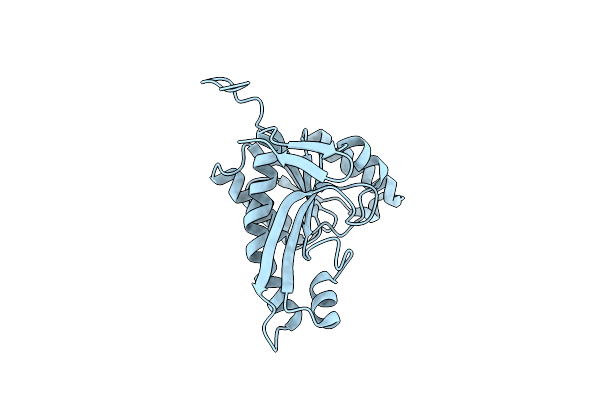
Crystal Structure Of Formyltetrahydrofolate Deformylase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-01-08 Classification: HYDROLASE |
 |
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Symbiobacterium Toebii
Organism: Symbiobacterium toebii
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2012-03-07 Classification: TRANSFERASE |
 |
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Geobacillus Kaustophilus
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-03-07 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2007-10-23 Classification: TRANSFERASE Ligands: MG, CO |

