Search Count: 71
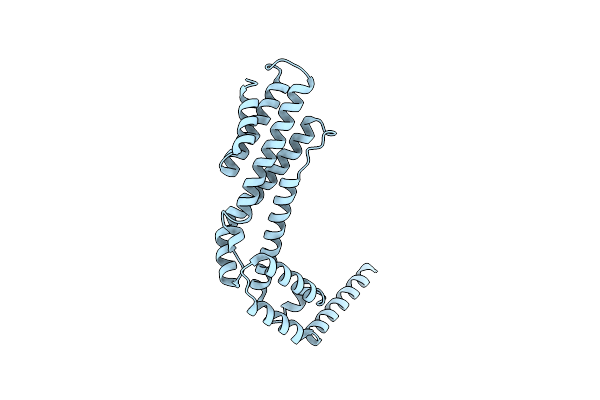 |
Crystal Structure Of The C-Terminal Fragment (Residues 756-982 With The C864S Mutation) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
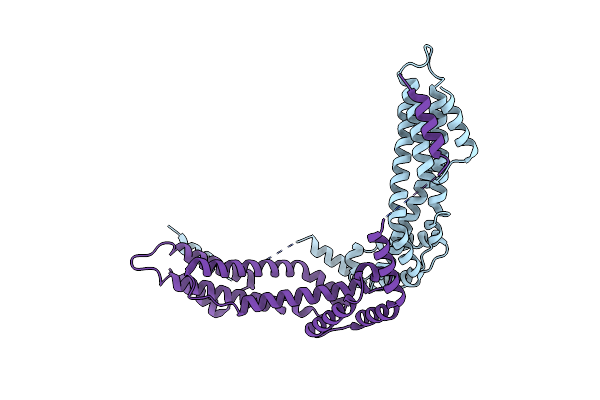 |
Crystal Structure Of The C-Terminal Fragment (Residues 716-982) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, NA, CYN, TGL, PGV, EDO, CUA, CHD, PEK, CDL, DMU, PSC, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In The Cyanide-Bound Fully Oxidized State At 50 K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CYN, PGV, EDO, TGL, CUA, PSC, CHD, PEK, CDL, DMU, ZN, PO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-11-16 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CYN, TGL, PGV, EDO, CUA, PSC, CHD, PEK, CDL, DMU, ZN, PO4 |
 |
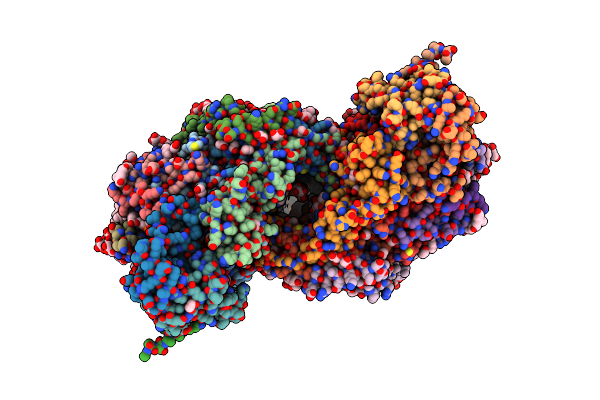
The Structure Of Azide-Bound Cytochrome C Oxidase Determined Using The Crystals Exposed To 20 Mm Azide Solution For 4 Days
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, AZI, PGV, EDO, TGL, CHD, CUA, PSC, DMU, CDL, PEK, ZN |
 |
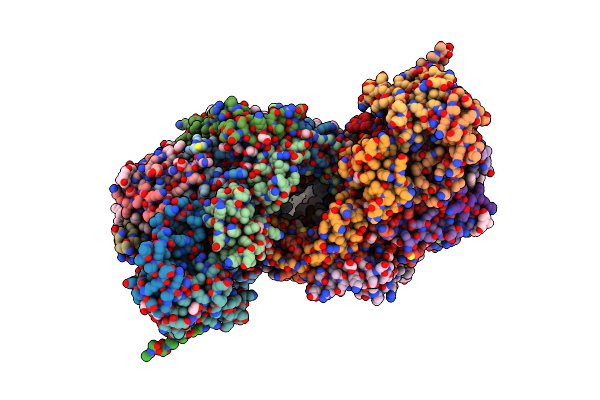
Bovine Heart Cytochrome C Oxidase In Fully Oxidized State At 1.5 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, EDO, DMU, CUA, CHD, PSC, CDL, PEK, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate Of E At 1.76 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, PSC, PER, EDO, TGL, CUA, CHD, DMU, CDL, PEK, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate Of O At 1.84 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, TGL, PGV, PER, PSC, EDO, DMU, CUA, CHD, PEK, CDL, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In A Catalytic Intermediate, Io10, At 1.74 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, OXY, EDO, DMU, TGL, CUA, PSC, CHD, PEK, CDL, ZN, PO4 |
 |
Bovine Heart Cytochrome C Oxidase In The Carbon Monoxide-Bound Fully Reduced State At A 50 K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: OXIDOREDUCTASE Ligands: PGV, HEA, CU, MG, NA, CMO, EDO, TGL, PSC, CHD, CUA, PEK, CDL, DMU, ZN, PO4 |
 |
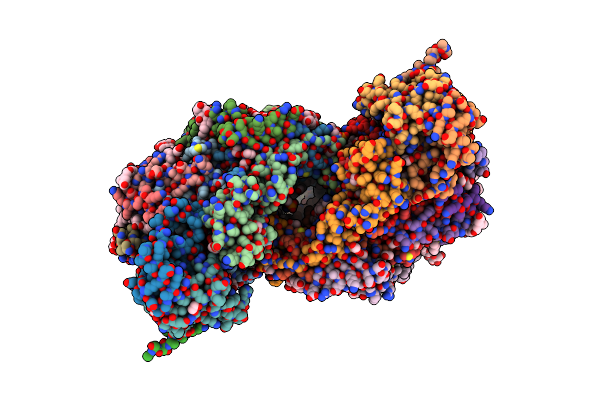
Binary Complex Of 14-3-3 Sigma And A High-Affinity Non-Canonical 9-Mer Peptide Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-01 Classification: PEPTIDE BINDING PROTEIN Ligands: MG, CL |
 |
Bovine Heart Cytochrome C Oxidase In Catalitic Intermediates At 1.80 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, DMU, PGV, PSC, TGL, EDO, CUA, CHD, CDL, PEK, ZN, PO4 |
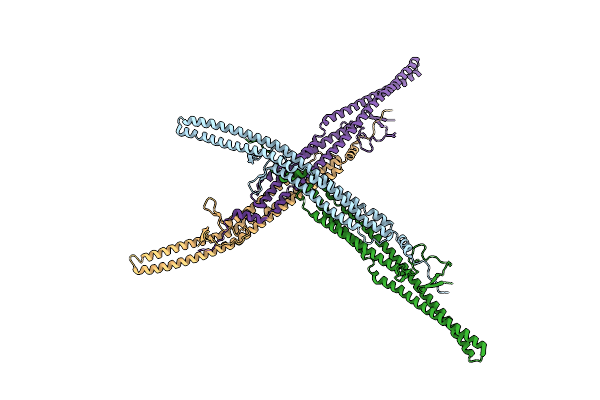 |
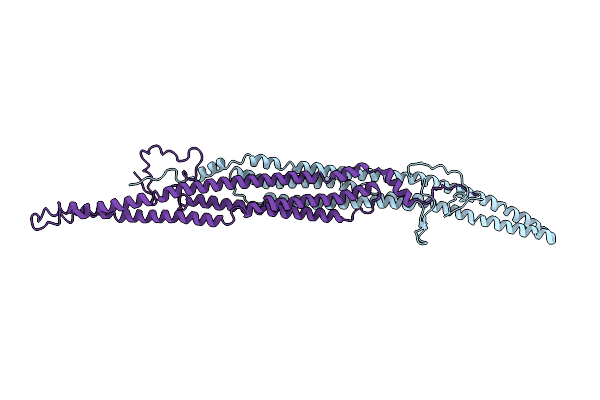
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN |
 |
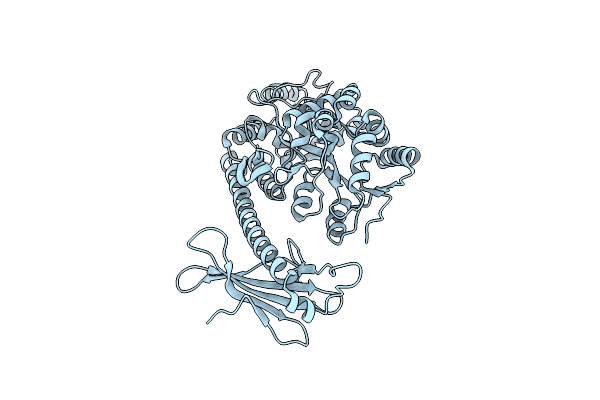
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.76 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN |
 |
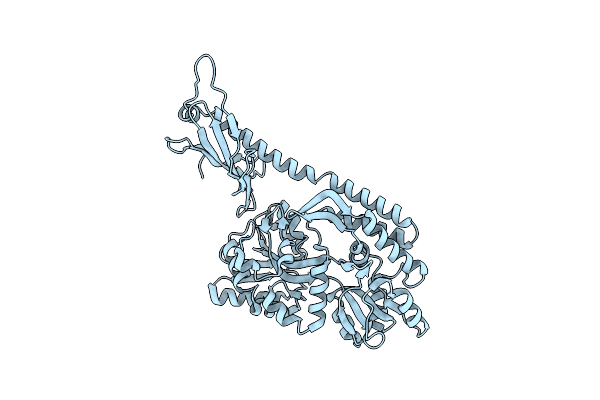
Crystal Structure Of Mbpapo-Tim21 Fusion Protein With A 16-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
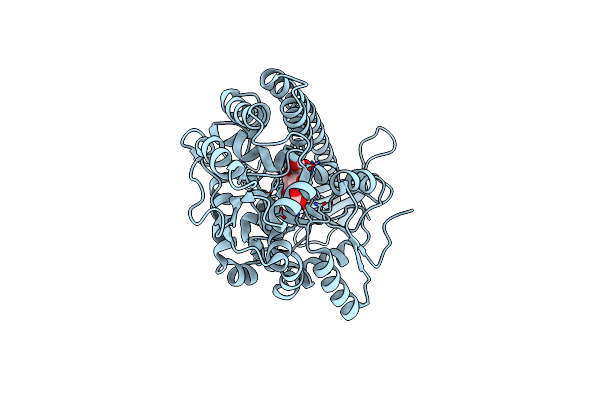
Crystal Structure Of Mbpapo-Tim21 Fusion Protein With A 17-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
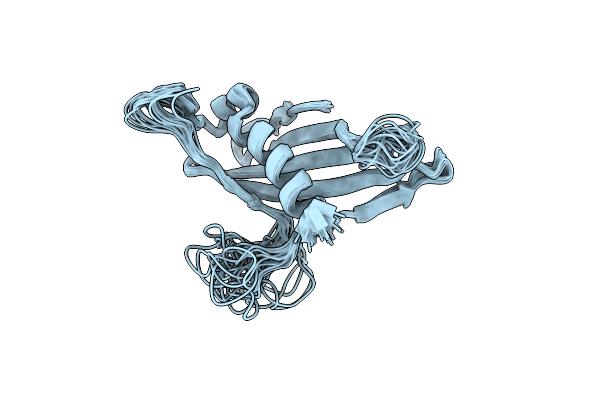
Crystal Structure Of Mbpholo-Tim21 Fusion Protein With A 17-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
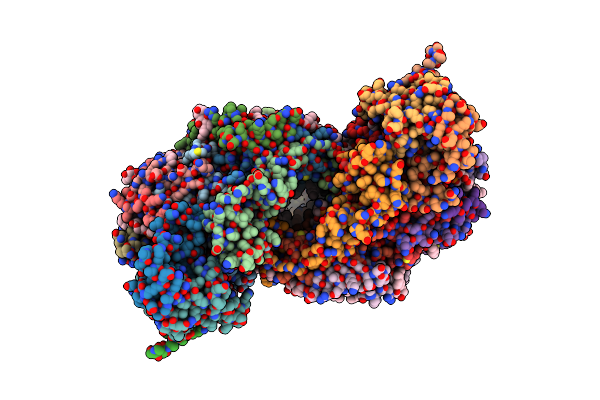
Solution Structure Of The Intermembrane Space Domain Of The Mitochondrial Import Protein Tim21 From S. Cerevisiae
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2019-09-11 Classification: PROTEIN TRANSPORT |
 |
Low-Dose Structure Of Bovine Heart Cytochrome C Oxidase In The Fully Oxidized State Determined Using 30 Kev X-Ray
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PGV, PER, TGL, CUA, CHD, PEK, CDL, DMU, PSC, ZN |

