Search Count: 16
 |
Duf2436 Domain Which Is Frequently Found In Virulence Proteins From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2024-10-09 Classification: UNKNOWN FUNCTION |
 |
Synthesis And Evaluation Of Diaryl Ether Modulators Of The Leukotriene A4 Hydrolase Aminopeptidase Activity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-08-28 Classification: HYDROLASE Ligands: VI7, ZN |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-08-07 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aspartate Semialdehyde Dehydrogenase From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aspartate Semialdehyde Dehydrogenase From Porphyromonas Gingivalis Complexed With 2',5'Adenosine Diphosphate
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: A2P |
 |
Organism: Burkholderiaceae
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-01-10 Classification: HYDROLASE |
 |
Substrate-Dependent Divergence Of Leukotriene A4 Hydrolase Aminopeptidase Activity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-06-22 Classification: HYDROLASE Ligands: ZN, N0Y, 28T |
 |
Substrate-Dependent Divergence Of Leukotriene A4 Hydrolase Aminopeptidase Activity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-06-15 Classification: HYDROLASE Ligands: ZN, N0Y, PGE |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-06-01 Classification: IMMUNE SYSTEM |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: NUCLEAR PROTEIN/Transferase Ligands: ZN, SF4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: NUCLEAR PROTEIN/DNA Ligands: CA, SF4, ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: NUCLEAR PROTEIN/DNA Ligands: SF4, ZN, CA |
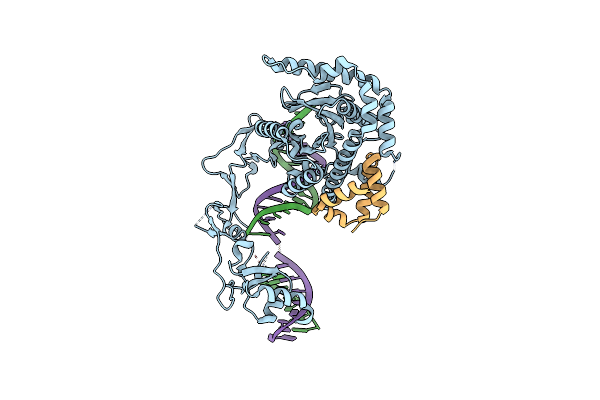 |
Role Of Beta-Hairpin Motifs In The Dna Duplex Opening By The Rad4/Xpc Nucleotide Excision Repair Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2020-10-14 Classification: DNA BINDING PROTEIN/DNA |
 |
The Effect Of Modifier Structure On The Activation Of Leukotriene A4 Hydrolase Aminopeptidase Activity.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-12-04 Classification: HYDROLASE Ligands: LNJ, ZN |
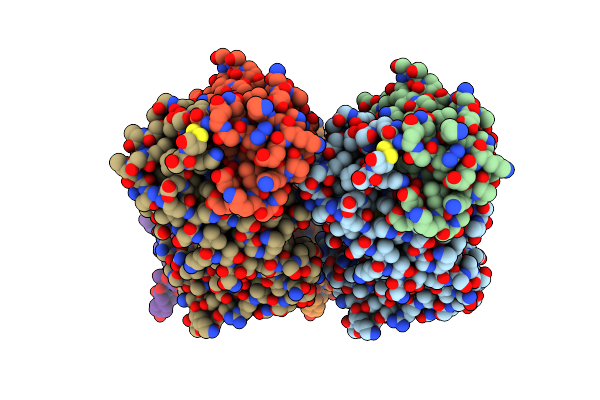 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2017-06-07 Classification: IMMUNE SYSTEM Ligands: A2G |
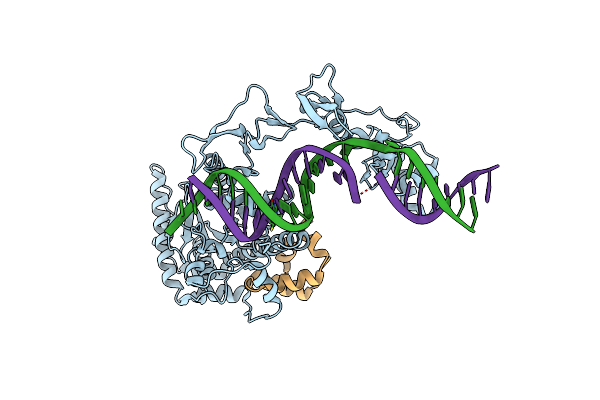 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2015-03-11 Classification: DNA BINDING PROTEIN/DNA |

