Search Count: 15
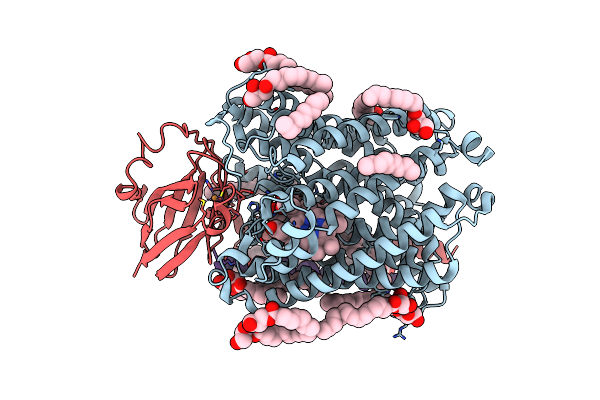 |
Serial Synchrotron Crystallography Structure Of Ba3-Type Cytochrome C Oxidase From Thermus Thermophilus Using A Goniometer Compatible Flow-Cell
Organism: Thermus thermophilus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-03-01 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CUA |
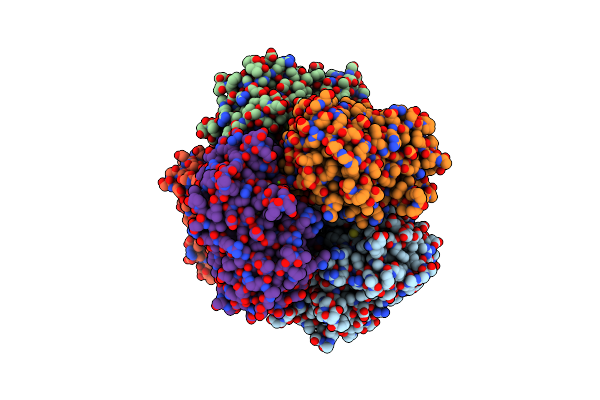 |
Sfx Structure Of Dye-Type Peroxidase Dtpb D152A Variant In The Ferryl State
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
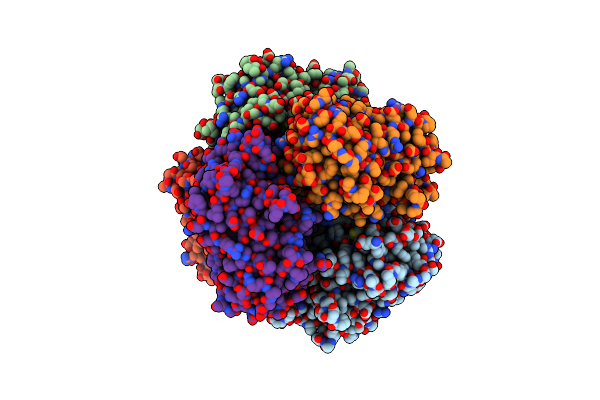 |
Sfx Structure Of Dye-Type Peroxidase Dtpb D152A/N245A Variant In The Ferric State
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HEM, MG |
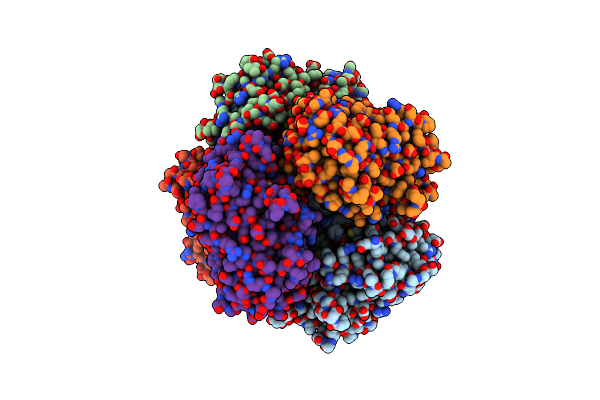 |
Sfx Structure Of Dye-Type Peroxidase Dtpb N245A Variant In The Ferric State
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HEM, MG |
 |
Sfx Structure Of Dye-Type Peroxidase Dtpb D152A Variant In The Ferric State
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HEM, MG |
 |
Sfx Structure Of Dye-Type Peroxidase Dtpb R243A Variant In The Ferric State
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HEM, MG |
 |
Crystal Structure Of Ribonucleotide Reductase R2 Subunit Solved By Serial Synchrotron Crystallography
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: MN3, FE |
 |
Structure Of Galectin-3C In Complex With Lactose Determined By Serial Crystallography Using A Silicon Nitride Membrane Support
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN |
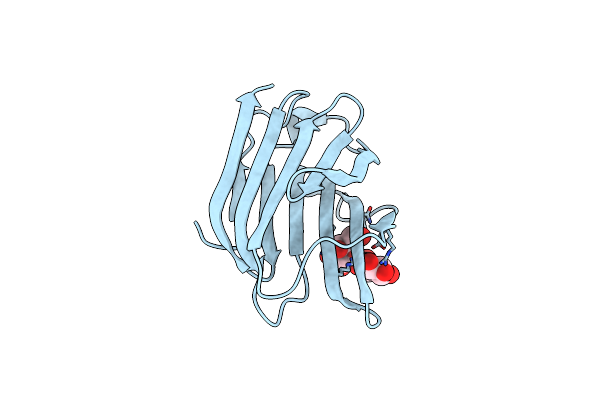 |
Structure Of Galectin-3C In Complex With Lactose Determined By Serial Crystallography Using An Xtaltool Support
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-17 Classification: SUGAR BINDING PROTEIN Ligands: CL |
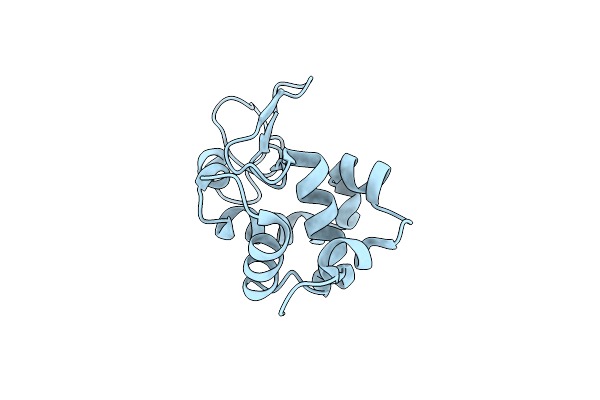 |
Room Temperature Crystal Structure Of Lysozyme Determined By Serial Synchrotron Crystallography (Micro Focused Beam - Crystfel)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-05-06 Classification: HYDROLASE Ligands: CL |
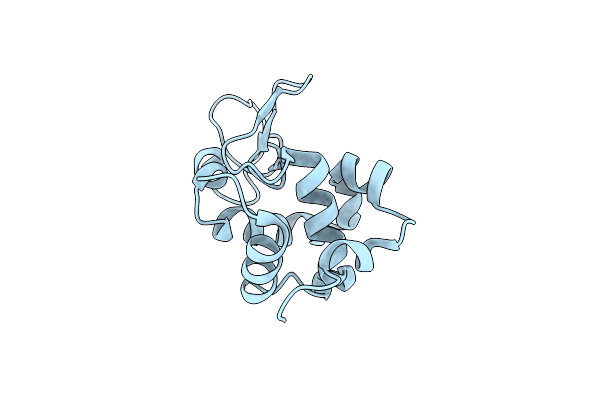 |
Room-Temperature Crystal Structure Of Lysozyme Determined By Serial Synchrotron Crystallography Using A Nano Focused Beam.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-05-06 Classification: HYDROLASE Ligands: CL |
 |
Raster-Scanning Protein Crystallography Using Micro And Nano-Focused Synchrotron Beams
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-05-06 Classification: HYDROLASE Ligands: CL |
 |
Room-Temperature Crystal Structure Of Lysozyme Determined By Serial Synchrotron Crystallography Using A Micro Focused Beam (Conventional Resolution Cut-Off)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-05-06 Classification: HYDROLASE Ligands: CL |
 |
Room Temperature Structure Of Bacteriorhodopsin From Lipidic Cubic Phase Obtained With Serial Millisecond Crystallography Using Synchrotron Radiation
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-02-18 Classification: PROTON TRANSPORT Ligands: RET, LI1 |
 |
Bacteriorhodopsin Ground State Structure Collected In Cryo Conditions From Crystals Obtained In Lcp With Peg As A Precipitant.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-02-18 Classification: PROTON TRANSPORT Ligands: RET, LI1 |

